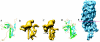Common ancestry of herpesviruses and tailed DNA bacteriophages - PubMed (original) (raw)
Common ancestry of herpesviruses and tailed DNA bacteriophages
Matthew L Baker et al. J Virol. 2005 Dec.
Abstract
Comparative analysis of capsid protein structures in the eukaryote-infecting herpesviruses (Herpesviridae) and the prokaryote-infecting tailed DNA bacteriophages (Caudovirales) revealed a characteristic fold that is restricted to these two virus lineages and is indicative of common ancestry. This fold not only serves as a major architectural element in capsid stability but also enables the conformational flexibility observed during viral assembly and maturation. On the basis of this and other emerging relationships, it seems increasingly likely that the very diverse collection of extant viruses may have arisen from a relatively small number of primordial progenitors.
Figures
FIG. 1.
A gallery of bacteriophage capsid protein structures determined by either X-ray crystallography or cryoEM. HK97 gp5 (A), mature P22 gp5 (B), procapsid P22 gp5 (C), and T4 gp24 (D) are shown in comparison to HSV-1 VP5 (E). VP5, the 145-kDa capsid protein, was segmented from an approximately 8-Å cryoEM map of the HSV-1 capsid. The red line demarcates the boundary between the floor domain and the other two domains of VP5 (upper and middle domains). The N-terminal helix in P22 that has been proposed to undergo refolding is indicated by the arrow in panel C.
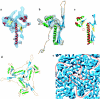
FIG. 2.
Match of the secondary structure elements of HSV-1 capsid VP5 and HK97 phage gp5 and their molecular interactions in the capsids. (a) The isolated VP5 floor domain, in blue, viewed from outside the capsid. SSEhunter identified two long α-helices (red; α1 and α2) adjacent to a large β-sheet (yellow; β1) in the floor domain, as well as a second β-sheet (yellow; β2) and several smaller helices flanking α1 and α2. (b) The HK97 capsid protein (gp5) shown in the same view reveals a similar structural motif. A simulated density map for gp5 at approximately 8-Å resolution is shown in pale blue. (c) Alignment of the secondary structure elements by use of Foldhunter (12) demonstrated a clear match between the floor domain of VP5 and the core structure of gp5. (d) Arrangement of gp5 subunits around a local three-fold axis (▴) in the HK97 capsid as viewed from inside the capsid. (e) Organization of the HSV-1 capsid floor as shown in the same view in panel d. Individual VP5 subunit floor domains are demarcated with the long α-helices (α1, α2) and associated β-sheets (β1, β2) annotated in one subunit.
Similar articles
- Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria.
Krupovic M, Forterre P, Bamford DH. Krupovic M, et al. J Mol Biol. 2010 Mar 19;397(1):144-60. doi: 10.1016/j.jmb.2010.01.037. Epub 2010 Jan 25. J Mol Biol. 2010. PMID: 20109464 - Tailed bacteriophages: the order caudovirales.
Ackermann HW. Ackermann HW. Adv Virus Res. 1998;51:135-201. doi: 10.1016/s0065-3527(08)60785-x. Adv Virus Res. 1998. PMID: 9891587 Free PMC article. Review. - Diversity of temperate bacteriophages induced in bovine and ovine Mannheimia haemolytica isolates and identification of a new P2-like phage.
Davies RL, Lee I. Davies RL, et al. FEMS Microbiol Lett. 2006 Jul;260(2):162-70. doi: 10.1111/j.1574-6968.2006.00314.x. FEMS Microbiol Lett. 2006. PMID: 16842340 - Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy.
Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Jiang W, et al. Nature. 2008 Feb 28;451(7182):1130-4. doi: 10.1038/nature06665. Nature. 2008. PMID: 18305544 - Herpesvirus assembly: an update.
Mettenleiter TC, Klupp BG, Granzow H. Mettenleiter TC, et al. Virus Res. 2009 Aug;143(2):222-34. doi: 10.1016/j.virusres.2009.03.018. Epub 2009 Apr 7. Virus Res. 2009. PMID: 19651457 Review.
Cited by
- Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems.
Fokine A, Rossmann MG. Fokine A, et al. Structure. 2016 Nov 1;24(11):1928-1935. doi: 10.1016/j.str.2016.08.013. Epub 2016 Sep 22. Structure. 2016. PMID: 27667692 Free PMC article. - Reconstructing virus structures from nanometer to near-atomic resolutions with cryo-electron microscopy and tomography.
Chang J, Liu X, Rochat RH, Baker ML, Chiu W. Chang J, et al. Adv Exp Med Biol. 2012;726:49-90. doi: 10.1007/978-1-4614-0980-9_4. Adv Exp Med Biol. 2012. PMID: 22297510 Free PMC article. Review. - Encapsulins-Bacterial Protein Nanocompartments: Structure, Properties, and Application.
Gabashvili AN, Chmelyuk NS, Efremova MV, Malinovskaya JA, Semkina AS, Abakumov MA. Gabashvili AN, et al. Biomolecules. 2020 Jun 26;10(6):966. doi: 10.3390/biom10060966. Biomolecules. 2020. PMID: 32604934 Free PMC article. Review. - Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus.
Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Jiang W, et al. Nature. 2006 Feb 2;439(7076):612-6. doi: 10.1038/nature04487. Nature. 2006. PMID: 16452981 Free PMC article. - Three-dimensional structure of tropism-switching Bordetella bacteriophage.
Dai W, Hodes A, Hui WH, Gingery M, Miller JF, Zhou ZH. Dai W, et al. Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4347-52. doi: 10.1073/pnas.0915008107. Epub 2010 Feb 16. Proc Natl Acad Sci U S A. 2010. PMID: 20160083 Free PMC article.
References
- Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. - PubMed
- Casjens, S., and J. King. 1975. Virus assembly. Annu. Rev. Biochem. 44:555-611. - PubMed
- Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. - PubMed
- Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. - PubMed
- Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources