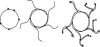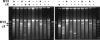Cell-free cloning using phi29 DNA polymerase - PubMed (original) (raw)
Comparative Study
. 2005 Nov 29;102(48):17332-6.
doi: 10.1073/pnas.0508809102. Epub 2005 Nov 14.
Affiliations
- PMID: 16286637
- PMCID: PMC1283157
- DOI: 10.1073/pnas.0508809102
Comparative Study
Cell-free cloning using phi29 DNA polymerase
Clyde A Hutchison 3rd et al. Proc Natl Acad Sci U S A. 2005.
Abstract
We describe conditions for rolling-circle amplification (RCA) of individual DNA molecules 5-7 kb in size by >10(9)-fold, using phi29 DNA polymerase. The principal difficulty with amplification of small amounts of template by RCA using phi29 DNA polymerase is "background" DNA synthesis that usually occurs when template is omitted, or at low template concentrations. Reducing the reaction volume while keeping the amount of template fixed increases the template concentration, resulting in a suppression of background synthesis. Cell-free cloning of single circular molecules by using phi29 DNA polymerase was achieved by carrying out the amplification reactions in very small volumes, typically 600 nl. This procedure allows cell-free cloning of individual synthetic DNA molecules that cannot be cloned in Escherichia coli, for example synthetic phage genomes carrying lethal mutations. It also allows cell-free cloning of genomic DNA isolated from bacteria. This DNA can be sequenced directly from the phi29 DNA polymerase reaction without further amplification. In contrast to PCR amplification, RCA using phi29 DNA polymerase does not produce mutant jackpots, and the high processivity of the enzyme eliminates stuttering at homopolymer tracts. Cell-free cloning has many potential applications to both natural and synthetic DNA. These include environmental DNA samples that have proven difficult to clone and synthetic genes encoding toxic products. The method may also speed genome sequencing by eliminating the need for biological cloning.
Figures
Fig. 1.
RCA by φ29 DNA polymerase, after Dean et al. (5). Arrowheads represent random hexamer primers. This drawing is not to scale; circular templates are typically 5 kb, and product strands average 70 kb in length.
Fig. 2.
Factors affecting background synthesis by φ29 polymerase. (A) Background increases with decreasing template concentration. Three-microliter φ29 polymerase reactions were primed by the indicated numbers of φX single-stranded DNA molecules, calculated on the basis of dilution factors from a stock of known concentration. Reactions were digested with PstI and analyzed as described above. (B) Background synthesis is suppressed by reducing φ29 polymerase reaction volume. φ29 polymerase reactions of 30, 15, and 3 μl were assembled, each containing either 50 M13 single-stranded DNA molecules or no template DNA. The reaction products were digested with PstI, and one-half of each reaction was analyzed by gel electrophoresis as described above. The products of background synthesis, not cleaved by PstI, migrate as large DNA, slower than the largest marker DNA (10 kb).
Fig. 3.
Cell-free cloning of φX174 and M13 from a mixture. Twenty duplicate 600-nl φ29 polymerase reactions were made by using the same limiting dilution of a mixture of φX174 and M13 DNAs (see text). The reaction products were digested with PstI and analyzed as described above. The + and – symbols above the gel indicate the presence or absence of bands with the molecular weights of linear φX174 (5.4 kb) or M13 (7.2 kb) DNAs. The products of background synthesis, not cleaved by PstI, migrate as large DNA, slower than the largest marker DNA (10 kb).
Fig. 4.
DNA sequencing of φ29 polymerase clones. (A) Sequencing of cell-free clones of synthetic φX174 molecules. Sequencing was performed after PCR amplification of single-molecule φ29 polymerase reactions. The same region is compared for a synthetic φX clone with a single base deletion (molecule 1), one with the wild-type sequence in this region (molecule 3), and natural φX DNA. Signal strengths were as follows: molecule 1, A = 602, C = 607, G = 512, and T = 459; molecule 3, A = 324, C = 331, G = 292, and T = 300; natural φX, A = 1,242, C = 1,430, G = 1,199, and T = 1,154. (B) Direct sequencing of φ29 polymerase reaction products without PCR amplification. This figure shows the result of sequencing 18% of the product from a 600-nl reaction as described in Materials and Methods. Signal strengths were A = 48, C = 25, G = 29, and T = 52. (C) Sequence reads through an A18 tract and the complementary T18 tract on a molecule amplified by φ29 polymerase. Sequencing was performed on 18% of a 600-nl φ29 polymerase reaction, without PCR amplification. Primers, designed on the basis of the M. genitalium genome sequence, were 5′-GTTAAAGGGCGACTAATAG-3′ for the A18 sequence and 5′-AACTTAATACTTTGGTCAG-3′ for the T18 sequence. Signal strengths were A = 202, C = 104, G = 120, and T = 226 for the A18 sequence and A = 107, C = 57, G = 65, and T = 121 for the T18 sequence.
Fig. 5.
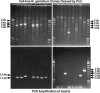
Cell-free cloning of M. genitalium genomic DNA. A library consisting of 1.5- to 2.5-kb fragments of M. genitalium DNA ligated to a pBR322 derivative was diluted 100-fold in TE from an initial DNA concentration of ≈0.3 ng/μl, heated at 95°C for 2 min, and quenched on ice. The DNA was diluted an additional 1,000-fold (Upper Left) or 2,000-fold (Upper Right) and used as template in two sets of 600-nl φ29 polymerase reactions (eight duplicate reactions per set), as described in Materials and Methods. One-half of each reaction was cleaved with PstI and analyzed by gel electrophoresis (Upper). Inserts were amplified by PCR from the remaining portion of each reaction, using M13 forward and reverse sequencing primers that flank the cloning site in the vector, and analyzed on the lower gels.
Similar articles
- Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification.
Dean FB, Nelson JR, Giesler TL, Lasken RS. Dean FB, et al. Genome Res. 2001 Jun;11(6):1095-9. doi: 10.1101/gr.180501. Genome Res. 2001. PMID: 11381035 Free PMC article. - A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase.
Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T. Inoue-Nagata AK, et al. J Virol Methods. 2004 Mar 15;116(2):209-11. doi: 10.1016/j.jviromet.2003.11.015. J Virol Methods. 2004. PMID: 14738990 - Optimal DNA templates for rolling circle amplification revealed by in vitro selection.
Mao Y, Liu M, Tram K, Gu J, Salena BJ, Jiang Y, Li Y. Mao Y, et al. Chemistry. 2015 May 26;21(22):8069-74. doi: 10.1002/chem.201500994. Epub 2015 Apr 15. Chemistry. 2015. PMID: 25877998 - Rolling circle amplification (RCA)-based DNA hydrogel.
Yao C, Zhang R, Tang J, Yang D. Yao C, et al. Nat Protoc. 2021 Dec;16(12):5460-5483. doi: 10.1038/s41596-021-00621-2. Epub 2021 Oct 29. Nat Protoc. 2021. PMID: 34716450 Review. - Rolling-circle amplification of viral DNA genomes using phi29 polymerase.
Johne R, Müller H, Rector A, van Ranst M, Stevens H. Johne R, et al. Trends Microbiol. 2009 May;17(5):205-11. doi: 10.1016/j.tim.2009.02.004. Epub 2009 Apr 15. Trends Microbiol. 2009. PMID: 19375325 Review.
Cited by
- A sensitive one-pot ROA assay for rapid miRNA detection.
Hou Z, Deng W, Li A, Zhang Y, Chang J, Guan X, Chang Y, Wang K, Wang X, Ruan J. Hou Z, et al. aBIOTECH. 2024 Mar 18;5(3):298-308. doi: 10.1007/s42994-024-00140-0. eCollection 2024 Sep. aBIOTECH. 2024. PMID: 39279850 Free PMC article. - scMicrobe PTA: near complete genomes from single bacterial cells.
Bowers RM, Gonzalez-Pena V, Wardhani K, Goudeau D, Blow MJ, Udwary D, Klein D, Vill AC, Brito IL, Woyke T, Malmstrom RR, Gawad C. Bowers RM, et al. ISME Commun. 2024 Jul 12;4(1):ycae085. doi: 10.1093/ismeco/ycae085. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39021442 Free PMC article. - A primer-independent DNA polymerase-based method for competent whole-genome amplification of intermediate to high GC sequences.
Ordóñez CD, Mayoral-Campos C, Egas C, Redrejo-Rodríguez M. Ordóñez CD, et al. NAR Genom Bioinform. 2023 Aug 21;5(3):lqad073. doi: 10.1093/nargab/lqad073. eCollection 2023 Sep. NAR Genom Bioinform. 2023. PMID: 37608803 Free PMC article. - Back to Basics: A Simplified Improvement to Multiple Displacement Amplification for Microbial Single-Cell Genomics.
Sobol MS, Kaster AK. Sobol MS, et al. Int J Mol Sci. 2023 Feb 21;24(5):4270. doi: 10.3390/ijms24054270. Int J Mol Sci. 2023. PMID: 36901710 Free PMC article.
References
- Esteban, J. A., Salas, M. & Blanco, L. (1993) J. Biol. Chem. 268, 2719–2726. - PubMed
- Garmendia, C., Bernad, A., Esteban, J. A., Blanco, L. & Salas, M. (1992) J. Biol. Chem. 267, 2594–2599. - PubMed
- Blanco, L., Bernad, A., Lazaro, J. M., Martin, G., Garmendia, C. & Salas, M. (1989) J. Biol. Chem. 264, 8935–8940. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources