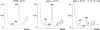Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes - PubMed (original) (raw)
Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes
Tiffany D Miles et al. Mol Cell Biol. 2005 Dec.
Abstract
The essential, conserved yeast nucleolar protein Ytm1 is one of 17 proteins in ribosome assembly intermediates that contain WD40 protein-protein interaction motifs. Such proteins may play key roles in organizing other molecules necessary for ribosome biogenesis. Ytm1 is present in four consecutive 66S preribosomes containing 27SA2, 27SA3, 27SB, and 25.5S plus 7S pre-rRNAs plus ribosome assembly factors and ribosomal proteins. Ytm1 binds directly to Erb1 and is present in a heterotrimeric subcomplex together with Erb1 and Nop7, both within preribosomes and independently of preribosomes. However, Nop7 and Erb1 assemble into preribosomes prior to Ytm1. Mutations in the WD40 motifs of Ytm1 disrupt binding to Erb1, destabilize the heterotrimer, and delay pre-rRNA processing and nuclear export of preribosomes. Nevertheless, 66S preribosomes lacking Ytm1 remain otherwise intact.
Figures
FIG. 4.
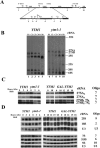
Processing of pre-rRNAs is altered in the _ytm1_-1 mutant. (A) Oligonucleotide probes or primers used to detect rRNAs and pre-rRNAs. (B) Yeast strains JWY3400 (YTM1) and JWY7128 (_ytm1_-1) were grown in YEPD medium at 25°C and shifted to 37°C for 5 h. Cells were pulse-labeled with [5,6 3H]uracil for 5 min and chased with an excess of unlabeled uracil for 2, 5, 10, and 60 min. Equal cpm of RNA isolated from cells at each time point were subjected to electrophoresis on agarose-formaldehyde gels to separate each pre-rRNA or rRNA and detected by autoradiography. (C) Primer extension was performed to determine steady-state levels of 27SA2, 27SA3, 27SBL plus 7SL, and 27SBS plus 7SS pre-rRNAs. RNA was extracted from whole-cell extracts from strains JWY3400 (YTM1) and JWY7128 (_ytm1_-1) grown in YEPD medium at 25°C or shifted from 25°C to 37°C for 3 h or 6 h or from strain JWY6149 (YTM1) or JWY6992 (GAL-YTM1) grown in galactose-containing medium and shifted to glucose-containing medium for 0, 10, 12, 15, or 18 h. (D) Northern blotting was used to determine steady-state levels of 25S, 18S, 5.8S, and 5S rRNA and 7S pre-rRNA. High-molecular-weight RNAs were subjected to electrophoresis on agarose-formaldehyde gels, whereas acrylamide-urea gels were used to separate low-molecular-weight RNAs. U3 snoRNA was used as a loading control. RNA was quantified by phosphorimaging and normalized to U3 snoRNA.
FIG. 1.
Pre-rRNA processing and pre-rRNP maturation pathway in Saccharomyces cerevisiae. (A) The 35S pre-rRNA contains sequences for mature 18S, 5.8S, and 25S rRNAs (represented as thick horizontal lines) along with additional internal and external spacer sequences (represented as thin horizontal lines). The 35S pre-rRNA is transcribed by RNA polymerase I and rapidly modified and processed to produce 33S pre-rRNA. Cleavage of 33S pre-rRNA at site A0 generates 32S pre-rRNA. The 20S and 27SA2 pre-rRNA processing intermediates are generated through internal cleavage of 32S pre-rRNA at the A2 site. Subsequent processing and cleavage of 20S and 27SA2 pre-rRNAs result in the production of the mature 18S, 25S, and 5.8 rRNAs, respectively. 5S rRNA is transcribed separately by RNA polymerase III. (B) Pre-rRNA processing occurs in preribosomal particles. The 35S primary transcript is found within the 90S pre-rRNP (dark gray circle). Cleavage at site A2 initiates subunit-specific maturation, generating the 43S and 66S pre-rRNPs (light gray circles). The 43S preribosome is exported to the cytoplasm, where final steps in 20S maturation take place. Multiple 66S preribosomes exist that contain each of the 27S or 25.5S plus 7S pre-rRNA processing intermediates. The mature 40S subunit contains 18S rRNA, whereas the 60S subunit contains 25S, 5.8S, and 5S rRNA (white circles).
FIG. 2.
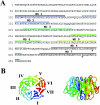
Ytm1 is a WD40 repeat-containing protein. (A) Predicted amino acid sequence of S. cerevisiae Ytm1. WD40 repeats are overlined. Amino acid residues altered in the _ytm1_-1 mutant are indicated by asterisks. (B) Ras Mol 2.6 was used to generate the top and side view of a model for amino acids 103 to 450 of Ytm1, based on the crystal structure of the WD repeat protein Gβ.
FIG. 3.
The _ytm1_-1 mutant is deficient in 60S ribosomal subunits. Free 40S and 60S ribosomal subunits, monoribosomes, and polyribosomes were assayed in yeast strains JWY3400 (YTM1) (left) or JWY7128 (_ytm1_-1) (center) grown at 25°C or JWY7128 grown at 25°C and shifted to 37°C for 3 h (right). Whole-cell extracts prepared from each strain were fractionated on 7 to 47% sucrose gradients. _A_260 peaks representing 40S and 60S ribosomal subunits and 80S monosomes are labeled. Half-mer polyribosomes are indicated by vertical arrows.
FIG. 5.
Inactivation of Ytm1 in the _ytm1_-1 mutant causes 66S preribosomes to accumulate in the nucleolus. The _ytm1_-1 mutant strain JWY6790 expressing eGFP-tagged rpL25 was grown in C-Trp medium at 25°C, washed and suspended in YEPD, and grown at 25°C (A and C) or shifted to 37°C for 5 h (B and D). Nuclei stained with 4′,6′-diamidino-2-phenylindole (DAPI) are shown in panels A and B (typically, nucleoli do not stain with DAPI). The signal from RpL25eGFP is shown in panels C and D. Arrows indicate nucleolar accumulation of rpL25eGFP (D) and corresponding DAPI staining (B).
FIG. 6.
Ytm1-HA3 cosediments on sucrose gradients with 66S preribosomes. Whole-cell extracts were prepared from yeast strain JWY6770 (YTM1-HA3) and fractionated on 7 to 47% sucrose velocity gradients. Fractions containing 40S and 60S ribosomal subunits and 80S monosomes are labeled. Proteins were trichloroacetic acid precipitated from gradient fractions and subjected to Western immunoblot analysis to detect Ytm1-HA3.
FIG. 7.
Ytm1 associates with pre-rRNAs in 66S preribosomes. (A) Whole-cell extracts were prepared from the YTM1-TAP strain JWY7124 and from untagged strain JWY3400 grown at 30°C in YEPD medium to 6 × 107 cells/ml. RNA was extracted from whole cells and from tandem affinity-purified samples, subjected to electrophoresis on agarose-formaldehyde or acrylamide-urea gels, blotted to nitrocellulose, and assayed by Northern blotting with specific oligonucleotide probes complementary to pre-rRNAs and mature rRNAs. Five percent of total RNA and 100% of tandem affinity-purified RNA were assayed. (B) Primer extension analysis was used to assay 35S, 27SA2, 27SA3, and 25.5S pre-rRNAs, as well as the BS and BL 5′ ends of 27S and 7S pre-rRNAs, using 32P-labeled oligonucleotides. Products of primer extension were resolved on sequencing gels, dried, and exposed to X-ray film for detection by autoradiography. (C) 35S pre-rRNA copurifies with TAP-tagged Erb1 and Nop7 but not Ytm1. RNA in whole-cell extracts and copurifying RNAs were assayed by primer extension as described above.
FIG. 8.
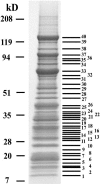
Nonribosomal proteins necessary for biogenesis of 60S ribosomal subunits, as well as ribosomal proteins, copurify with TAP-tagged Ytm1. Whole-cell extract was prepared from the YTM1-TAP strain JWY7124 grown at 30°C in YEPD medium to 6 × 107 cells/ml and subjected to tandem affinity purification. Proteins were trichloroacetic acid precipitated from column eluates and subjected to electrophoresis on 4 to 20% polyacrylamide gels. Proteins were stained with colloidal Coomassie blue, manually excised from the gel, digested with trypsin, and identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Table 2).
FIG. 9.
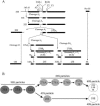
Ytm1, Erb1, and Nop7 form a heterotrimeric subcomplex both within 66S preribosomes and independently of these particles. (A) Ytm1, Erb1, and Nop7 are enriched (relative to other proteins found in 66S pre-rRNPs) among proteins copurifying with Ytm1-TAP or Nop7-TAP from _rrp1_-1 or _nop4_-3 mutants in which 66S preribosomes are unstable. Heterotrimer was purified (B) from sucrose gradient fractions, (C) by differential centrifugation, or (D) from whole-cell extracts treated with a cocktail of phosphatase inhibitors that disrupt 66S pre-rRNPs. Wild-type cells or mutant cells were grown at 25°C and shifted to 37°C for 5 h. Tandem affinity purification using Nop7-TAP or Ytm1-TAP was carried out from (A) whole-cell extracts from a 50-ml culture, (B) gradient fractions 5 to 7 (prepared from a 900-ml culture), (C) whole-cell extracts from 50 ml of cells (lanes 1 and 3) or 180,000 × g spin supernatants prepared from 500 ml of cells (lanes 2 and 4), or (D) untreated (−) or phosphatase inhibitor cocktail-treated extracts (+). Purified proteins were resolved by SDS-PAGE. Note that the heterotrimer is destabilized in the _ytm1_-1 mutant (B, lane 2; C, lane 4; D, lane 4). (E) Ytm1, Erb1, and Nop7 form a stable subcomplex within 66S preribosomes. Whole-cell extracts from YTM1 cells were subjected to centrifugation on 7 to 47% gradients. Fractions containing 66S preribosomes were pooled and subjected to tandem affinity purification in the presence (+) or absence (−) of phosphatase inhibitors, and proteins were resolved by SDS-PAGE. Bands indicated by asterisks in B and E are common contaminants that we observe upon TAP from any fractions of sucrose gradients (top, middle, or bottom) using any TAP-tagged protein.
FIG. 10.
Ytm1 and Nop7 directly interact with Erb1. (A) Synthetic radiolabeled proteins (*) were incubated with GST fusion proteins (lanes 2, 5, and 8). As negative controls, synthetic peptides were incubated with GST beads only (lanes 1, 4, and 7) or GST fusion proteins were incubated with the unrelated, radiolabeled 40S ribosome assembly factor Krr1 or ribosomal protein L11 (lanes 3, 6, and 9). (B) Radiolabeled wild-type Ytm1 protein was preincubated at 37°C for 15 min (lane 2). Mutant Ytm1-1 protein was preincubated at 37°C for 15 min (lane 3), 30 min (lane 4), or 60 min (lane 5). Following preincubation, wild-type Ytm1 or mutant Ytm1-1 radiolabeled protein was incubated with GST-Erb1. Mutant Ytm1-1 protein was incubated with GST beads only (lane 1) as a negative control. Complexes were eluted from glutathione beads, subjected to electrophoresis on 10% polyacrylamide gels, and detected by autoradiography. (C) Model for interactions between Ytm1, Erb1, and Nop7. Gray lines indicate interactions detected using GST pulldown assays, whereas the black line indicates interactions detected by two-hybrid assays.
FIG. 11.
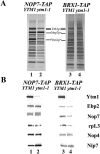
66S preribosomes are largely intact but lack Ytm1 in the _ytm1_-1 mutant. Wild-type YTM1 cells and mutant _ytm1_-1 cells expressing Nop7-TAP or Brx1-TAP were grown in YEPD medium at 25°C and shifted to 37°C for 5 h. (A) Silver staining or (B) Western immunoblot analysis was performed on proteins trichloroacetic acid precipitated from samples affinity purified from extracts from these strains.
Similar articles
- Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. Thoms M, et al. Nucleic Acids Res. 2016 Jan 29;44(2):926-39. doi: 10.1093/nar/gkv1365. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657628 Free PMC article. - Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes.
Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL Jr. Tang L, et al. Mol Biol Cell. 2008 Jul;19(7):2844-56. doi: 10.1091/mbc.e07-12-1281. Epub 2008 Apr 30. Mol Biol Cell. 2008. PMID: 18448671 Free PMC article. - The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14.
Granneman S, Nandineni MR, Baserga SJ. Granneman S, et al. Mol Cell Biol. 2005 Dec;25(23):10352-64. doi: 10.1128/MCB.25.23.10352-10364.2005. Mol Cell Biol. 2005. PMID: 16287850 Free PMC article. - Ribosome biogenesis in the yeast Saccharomyces cerevisiae.
Woolford JL Jr, Baserga SJ. Woolford JL Jr, et al. Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
- The human nucleolar protein FTSJ3 associates with NIP7 and functions in pre-rRNA processing.
Morello LG, Coltri PP, Quaresma AJ, Simabuco FM, Silva TC, Singh G, Nickerson JA, Oliveira CC, Moore MJ, Zanchin NI. Morello LG, et al. PLoS One. 2011;6(12):e29174. doi: 10.1371/journal.pone.0029174. Epub 2011 Dec 16. PLoS One. 2011. PMID: 22195017 Free PMC article. - Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis.
Rosado IV, Kressler D, de la Cruz J. Rosado IV, et al. Nucleic Acids Res. 2007;35(12):4203-13. doi: 10.1093/nar/gkm388. Epub 2007 Jun 13. Nucleic Acids Res. 2007. PMID: 17569673 Free PMC article. - Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing.
Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr. Sahasranaman A, et al. EMBO J. 2011 Sep 16;30(19):4020-32. doi: 10.1038/emboj.2011.338. EMBO J. 2011. PMID: 21926967 Free PMC article. - Nucleolar stress with and without p53.
James A, Wang Y, Raje H, Rosby R, DiMario P. James A, et al. Nucleus. 2014 Sep-Oct;5(5):402-26. doi: 10.4161/nucl.32235. Nucleus. 2014. PMID: 25482194 Free PMC article. Review. - Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I. Thoms M, et al. Nucleic Acids Res. 2016 Jan 29;44(2):926-39. doi: 10.1093/nar/gkv1365. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657628 Free PMC article.
References
- Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. L. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. - PubMed
- Baβler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
- Cagney, G., P. Uetz, and S. Fields. 2001. Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol. Genomics 7:27-34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F31 GM65067/GM/NIGMS NIH HHS/United States
- F31 GM19937/GM/NIGMS NIH HHS/United States
- F31 GM065067/GM/NIGMS NIH HHS/United States
- F31 GM019937/GM/NIGMS NIH HHS/United States
- R01 GM028301/GM/NIGMS NIH HHS/United States
- R01 GM28301/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases