Transcription repressor activity of spleen tyrosine kinase mediates breast tumor suppression - PubMed (original) (raw)
Transcription repressor activity of spleen tyrosine kinase mediates breast tumor suppression
Lei Wang et al. Cancer Res. 2005.
Abstract
Spleen tyrosine kinase (SYK) is a candidate tumor suppressor gene in breast. Loss of SYK expression in breast tumors as a result of DNA hypermethylation promotes tumor cell proliferation and invasion and predicts shorter survival of breast cancer patients. We previously reported that, in addition to its well-known cytoplasmic localization, the full-length Syk is also present in the nucleus and that Syk nuclear translocation is a rate-limiting step to determine Syk tumor suppressor function. Here, we show that the full-length form of Syk acts as a transcription repressor in the cell nucleus. Ectopic expression of Syk down-regulates the transcription of FRA1 and cyclin D1 oncogenes. This transcription-repressing activity of Syk is associated with its binding to members of the histone deacetylase family. Syk interacts with transcription factor Sp1 at the Sp1 DNA-binding site in the FRA1 promoter to repress Sp1-activated FRA1 transcription. Thus, breast tumorigenesis and progression resulting from the loss of SYK are underscored by the derepression of Sp1-mediated oncogene transcription.
Figures
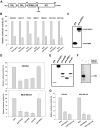
Figure 1.
Transcription repressor activity of Syk(L). A, The functional domains of the 653-amino-acid human Syk(L) protein are represented. K402 indicates the residue critical for the catalytic activity of Syk(L). B, HEK293, NIH3T3, MDA-MB-231, MDA-MB-435S, MCF10A, and MCF12A cells were transfected with pFA-SYK(L) or parental vector pFA-CMV, in combination with pRF-Luc. pCMVβ was included to normalize transfection efficiency. Gal4 luciferase activity was set at 100%. All reporter data were obtained from at least three independent experiments and are presented as means ± SE. C, Expression of Gal4-Syk(L) fusion proteins in HEK293 cells transfected with pFA or pFA-SYK(L) as detected by immunoblotting with an antibody against the Gal4 DNA-binding domain. D, Cells were transfected with pRF-Luc in combination with Gal4 constructs as described in (A). Luciferase activity of pFA was set at 100%. E, Immunodetection of Gal4 fusion proteins in HEK293 cells. F, Inactivation of kinase activity by K402R mutation of Syk. The wild-type (WT) or mutant (KI) pFlag-SYK(L) expression vectors were transfected into COS7 cells. Flag-immunoprecipitated products were incubated with cdb3 in an in vitro kinase reaction in the presence of [γ-32P]ATP. Piceatannol (100 nM) was used to control the specificity of kinase reactions. G, HEK293 and MDA-MB-231 cells were transfected with pRF-Luc and pCMVβ, together with pFA, pFA-SYK(L), or pFA-SYK(L)-KI. Reporter activity was determined as described in (A).
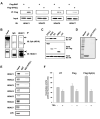
Figure 2.
In vivo and in vitro interactions of Syk(L) with HDACs. A, HEK293 cells were transfected with pFlag-SYK(L) or pFlag-BAP (bovine alkaline phosphatase; control). Two days following transfection, cells were lysed and subjected to Flag-immunoprecipitation (M2). The precipitated proteins were then probed with antibodies against HDACs 1, 3, 6, and 7. B, MDA-MB-468 cells were lysed followed by immunoprecipitation using an anti-HDAC1 antibody or rabbit IgG. The Syk protein (arrow) was detected in the precipitated materials by immunoblotting. Precipitation of HDAC1 (arrowhead) was verified by re-probing with the anti-HDAC1 antibody. Asterisks indicate the heavy chain of IgG. Input lysates were run side by side. C, pCMX-hHDAC1-HA was transfected into HEK293 cells, together with pFlag fusion proteins (pFlag-CMV was used as a control). The M2-precipitated products were measured for HA-immunoreactive substances by blotting. Input lysates were also analyzed for HA-HDAC1 and Flag-Syk expression by immunoblotting. D, Coomassie Blue staining of GST or GST-Syk(L) (arrow) fusion proteins. E, GST pull-down assays. HDACs 1-7 or an unrelated luciferase (LUC) protein were synthesized by TNT reactions in the presence of [35S]methionine. The labeled proteins were then incubated with immobilized GST or GST-Syk(L). Components bound to GST fusion were separated by SDS-PAGE and detected by autoradiography. Ten percent of the total input was run side by side to show relative binding capacity. F, HEK293 cells were transfected with pFlag-CMV (Flag), pFlag-SYK(L) [Syk(L)], or left untransfected (UT). Cell lysates were then M2-immunoprecipitated and used for HDAC activity assays. The TSA-containing reaction (5 μM) was used to determine the background fluorescence generated by non-HDAC signals. Data are means ± SE of four separate experiments.
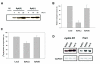
Figure. 3.
Expression of Syk(L), but not Syk(S), suppresses breast cancer cell invasiveness. A, Immunoblot detection of adenovirally delivered Syk in MDA-MB-231 cells. MOI, multiplicity of infection. B, Inhibition of cell invasion by adenovirally transduced Syk(L), but not Syk(S), as measured by chemoinvasion assays in MDA-MB-231 cells. The number of cells that invaded through Matrigel was compared to that in the LacZ group (arbitrarily set as 100%). Data are means ± SE from three experiments. C, Proliferation rate of MDA-MB-231 cells, as measured by [3H]thymidine incorporation, was inhibited by Syk(L) but not by Syk(S). D, Down-regulation of cyclin D1 and FRA1 mRNA by expression of Syk(L), but not Syk(S). Total RNA was extracted from MDA-MB-231 cells infected with Adeno-SYK(L), Adeno-SYK(S), or Adeno-LacZ. Northern blotting was done with random primer-labeled gene-specific cDNA products. The membrane was stripped and re-hybridized with GAPDH probe.
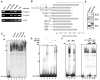
Figure. 4.
Interaction of Syk(L) with Sp1 at the Sp1-binding sequence of the FRA1 promoter. A, ChIP assays were performed using MDA-MB-231/Flag-Syk(L) stable cells. Two sets of FRA1 primers were used for PCR amplification from chromatin fragments immunoprecipitated by anti-Syk (4D10) and anti-Flag (M2) antibodies, mouse IgG (negative control), or anti-TFIIB antibody (positive control). Input represents PCR amplification of total chromatin before immunoprecipitation. A primer set of an unrelated region was used to normalize the amount of chromatin in each PCR reaction (bottom panel). B, MDA-MB-231/Flag-Syk(L) or its Neo control stable cells were transfected with the indicated reporter constructs. Bars represent means luciferase activity (± SE) relative to parental pGL3-basic (set at 100%). All luciferase measurements are from at least three independent experiments and normalized with activity in the Neo line. The relative positions of the putative Sp1 site and PCR primers (arrows) used for ChIP are shown. C, EMSA. 32P-labeled, 100-bp, Syk(L)-responsive fragment of the FRA1 promoter region was incubated with K562 cell nuclear extracts (NE) alone or in combination with anti-Syk antibodies (4D10, mouse monoclonal; or N19, rabbit polyclonal), anti-Sp1 antibody (rabbit polyclonal), or IgG controls. Ab, antibody. D, EMSA. The above-described DNA probe was incubated with recombinant Sp1 protein in combination with purified GST-Syk(L) or GST control protein in the presence or absence of Sp1. E, Lysates from HEK293 cells transfected with pFLAG-SYK(L) or pFLAG-BAP (control) were subjected to M2 immunoprecipitation and immunodetection of Sp1 (top panel). Lysate inputs were immunoblotted for Sp1 and Flag fusion proteins (center and bottom panels). F, Mutations at the putative Sp1 site disrupt DNA-protein complex formation. Left panel, EMSA was performed using K562 nuclear extracts (NE) and the 100-bp wild-type (WT) or Sp1-site-mutated (MUT) FRA1 promoter fragments as probes. Right panel, competition experiment using excess (10x and 100x) wild-type or mutant non-labeled probes.
Figure 5.
Comparison of Rb- and Syk-mediated transcription repression. Rb (or its family members p107 and p130) intereacts with chromatin remodeling enzymes such as HDACs and SWI/SNF. The interaction of Rb with E2F allows the remodeling enzymes to be targeted to promoters where they affect nucleosome assembly. Similarly, the interaction of Syk(L) with HDACs is responsible for Syk(L) transcription repression activity. The binding of Syk(L) to Sp1, which is either a direct interaction or through an intermediator factor (marked as “X”), predicts Syk's ability to govern Sp1-activated gene transcription.
Similar articles
- Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer.
Wang L, Duke L, Zhang PS, Arlinghaus RB, Symmans WF, Sahin A, Mendez R, Dai JL. Wang L, et al. Cancer Res. 2003 Aug 1;63(15):4724-30. Cancer Res. 2003. PMID: 12907655 - [Biological mechanism of full-length form of spleen tyrosine kinase regulating gene transcription in breast cancer cells].
Wang L, Hu Y, Yang ZL, Song XM, Wang JP. Wang L, et al. Ai Zheng. 2007 May;26(5):469-72. Ai Zheng. 2007. PMID: 17672934 Chinese. - [Spleen tyrosine kinase (L) suppresses breast cancer development by downregulating the expression of cyclin D1, ID1, B-myb and Fra1].
Wang L, Wang JP, Song XM, He YL. Wang L, et al. Zhonghua Yi Xue Za Zhi. 2007 Jan 9;87(2):85-9. Zhonghua Yi Xue Za Zhi. 2007. PMID: 17418011 Chinese. - Syk: a new player in the field of breast cancer.
Stewart ZA, Pietenpol JA. Stewart ZA, et al. Breast Cancer Res. 2001;3(1):5-7. doi: 10.1186/bcr261. Epub 2000 Nov 2. Breast Cancer Res. 2001. PMID: 11250739 Free PMC article. Review. - [Functions of spleen tyrosine kinase (Syk) gene and its correlation to neoplasms].
Wang HY, Zhang ZX. Wang HY, et al. Ai Zheng. 2007 May;26(5):555-60. Ai Zheng. 2007. PMID: 17672952 Review. Chinese.
Cited by
- Modulation by Syk of Bcl-2, calcium and the calpain-calpastatin proteolytic system in human breast cancer cells.
Fei B, Yu S, Geahlen RL. Fei B, et al. Biochim Biophys Acta. 2013 Oct;1833(10):2153-64. doi: 10.1016/j.bbamcr.2013.05.010. Epub 2013 May 16. Biochim Biophys Acta. 2013. PMID: 23684705 Free PMC article. - Migration inhibition of mammary epithelial cells by Syk is blocked in the presence of DDR1 receptors.
Neuhaus B, Bühren S, Böck B, Alves F, Vogel WF, Kiefer F. Neuhaus B, et al. Cell Mol Life Sci. 2011 Nov;68(22):3757-70. doi: 10.1007/s00018-011-0676-8. Epub 2011 Apr 17. Cell Mol Life Sci. 2011. PMID: 21499918 Free PMC article. - Phospho-proteomic analysis of mantle cell lymphoma cells suggests a pro-survival role of B-cell receptor signaling.
Pighi C, Gu TL, Dalai I, Barbi S, Parolini C, Bertolaso A, Pedron S, Parisi A, Ren J, Cecconi D, Chilosi M, Menestrina F, Zamò A. Pighi C, et al. Cell Oncol (Dordr). 2011 Apr;34(2):141-53. doi: 10.1007/s13402-011-0019-7. Epub 2011 Mar 11. Cell Oncol (Dordr). 2011. PMID: 21394647 Free PMC article. - The SYK tyrosine kinase: a crucial player in diverse biological functions.
Mócsai A, Ruland J, Tybulewicz VL. Mócsai A, et al. Nat Rev Immunol. 2010 Jun;10(6):387-402. doi: 10.1038/nri2765. Nat Rev Immunol. 2010. PMID: 20467426 Free PMC article. Review. - A PREVIOUSLY UNKNOWN UNIQUE CHALLENGE FOR INHIBITORS OF SYK ATP-BINDING SITE: ROLE OF SYK AS A CELL CYCLE CHECKPOINT REGULATOR.
Uckun FM, Ma H, Ozer Z, Goodman P, Zhang J, Qazi S. Uckun FM, et al. EBioMedicine. 2014 Nov 1;1(1):16-28. doi: 10.1016/j.ebiom.2014.10.019. EBioMedicine. 2014. PMID: 25506060 Free PMC article.
References
- Darnell JE. Transcription factors as targets for cancer therapy. Nature Rev Cancer. 2002;2:740–9. - PubMed
- Yuan YF, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61:5558–61. - PubMed
- Coopman PJ, Do MTH, Barth M, et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–7. - PubMed
- Toyama T, Iwase H, Yamashita H, et al. Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 2002;189:97–102. - PubMed
- Dejmek J, Leandersson K, Manjer J, et al. Expression and signaling activity of Wnt-5a/Discoidin Domain Receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin Cancer Res. 2005;11:520–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA100278/CA/NCI NIH HHS/United States
- R01 CA100278-03/CA/NCI NIH HHS/United States
- R01-CA100278/CA/NCI NIH HHS/United States
- R01-ES011863/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous