Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae - PubMed (original) (raw)
Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae
Toyoko Tsukuda et al. Nature. 2005.
Abstract
The repair of DNA double-strand breaks (DSBs) is crucial for maintaining genome stability. Eukaryotic cells repair DSBs by both non-homologous end joining and homologous recombination. How chromatin structure is altered in response to DSBs and how such alterations influence DSB repair processes are important issues. In vertebrates, phosphorylation of the histone variant H2A.X occurs rapidly after DSB formation, spreads over megabase chromatin domains, and is required for stable accumulation of repair proteins at damage foci. In Saccharomyces cerevisiae, phosphorylation of the two principal H2A species is also signalled by DSB formation, which spreads approximately 40 kb in either direction from the DSB. Here we show that near a DSB phosphorylation of H2A is followed by loss of histones H2B and H3 and increased sensitivity of chromatin to digestion by micrococcal nuclease; however, phosphorylation of H2A and nucleosome loss occur independently. The DNA damage sensor MRX is required for histone loss, which also depends on INO80, a nucleosome remodelling complex. The repair protein Rad51 (ref. 6) shows delayed recruitment to DSBs in the absence of histone loss, suggesting that MRX-dependent nucleosome remodelling regulates the accessibility of factors directly involved in DNA repair by homologous recombination. Thus, MRX may regulate two pathways of chromatin changes: nucleosome displacement for efficient recruitment of homologous recombination proteins; and phosphorylation of H2A, which modulates checkpoint responses to DNA damage.
Figures
Figure 1
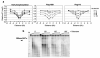
Chromatin changes at the MATαDSB.(a) A DSB was induced at _MAT_α in Flag-H2B or Flag-H3 expressing strains, and ChIP was performed with γ-H2A or anti-Flag antibodies. DNA was analyzed by real-time PCR using primers corresponding to sequences on the left (“−”) or right (“+”) side of the DSB (“0”), and results were normalized to the 0′ IP/Input DNA. Data in graphs are means +/− s.e.m. (b) Nuclei were prepared after DSB induction, and chromatin was digested with MNase and subjected to Southern blot analysis using a _MAT_α DNA probe. M=1 Kb DNA ladder.
Figure 2
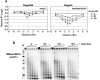
MRX plays a role in histone loss at the MATαDSB.(a) A DSB was induced at _MAT_α, and ChIP was performed with anti-Flag antibodies in an mre11::Kan-MX strain expressing Flag-H2B and an hta1/hta2-S129* strain expressing Flag-H3. DNA was analyzed by realtime PCR on both the left and right sides of the break site (“0”), and data are means +/− s.e.m.. (b) MNase analysis was performed on nuclei isolated from the mre11::Kan-MX strain by Southern blot analysis as described in Figure 1c.
Figure 3
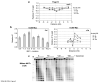
**The INO80 complex is required for histone eviction at the MAT**α DSB.(a) A DSB was induced at _MAT_α in an arp8::Kan-MX mutant expressing Flag-H3, and ChIP was performed with antibodies against the Flag epitope. Precipitated DNA was quantitated as described in Figure 1a. (b) ChIP was performed with anti-Myc antibodies in a wild type strain that contained Ino80-Myc before (−Gal) and after (+Gal) DSB induction. Ino80-Myc association was normalized to histone H3 occupancy. (c) MNase digestion was performed on nuclei isolated from an arp8::Kan-MX mutant as described in Figure 1c. Data in graphs are means +/− s.e.m.
Figure 4
**MRX and INO80 are required for recruitment of Rad51 to the MAT**α DSB.(a) ChIP was performed in wild type, _arp8_Δ, or _mre11_Δ with RPA (left) or Rad51 (right) antibodies after DSB induction at _MAT_α. DNA on the right side of the DSB was analyzed by real-time PCR, and data are means +/− s.e.m.. (b) MRX controls chromatin remodeling at DSBs. MRX is recruited to DSBs, regulating DNA end processing and Tel1-depenedent γ-H2A formation. MRX regulates nucleosome-remodelingthrough INO80, leading to nucleosome eviction and efficient recruitment of HR proteins. γ-H2A is independent of nucleosome displacement and controls accumulation of checkpoint proteins, as well as cohesin, at DSBs.
Similar articles
- The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks.
Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. Shim EY, et al. Mol Cell Biol. 2005 May;25(10):3934-44. doi: 10.1128/MCB.25.10.3934-3944.2005. Mol Cell Biol. 2005. PMID: 15870268 Free PMC article. - The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends.
Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. Chen X, et al. Nature. 2012 Sep 27;489(7417):576-80. doi: 10.1038/nature11355. Epub 2012 Sep 9. Nature. 2012. PMID: 22960743 Free PMC article. - The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast.
Wiest NE, Houghtaling S, Sanchez JC, Tomkinson AE, Osley MA. Wiest NE, et al. Nucleic Acids Res. 2017 Jun 2;45(10):5887-5900. doi: 10.1093/nar/gkx221. Nucleic Acids Res. 2017. PMID: 28398510 Free PMC article. - Nucleosome remodelers in double-strand break repair.
Seeber A, Hauer M, Gasser SM. Seeber A, et al. Curr Opin Genet Dev. 2013 Apr;23(2):174-84. doi: 10.1016/j.gde.2012.12.008. Epub 2013 Jan 23. Curr Opin Genet Dev. 2013. PMID: 23352131 Review. - Structure-function relationships of the Mre11 protein in the control of DNA end bridging and processing.
Marsella A, Cassani C, Casari E, Tisi R, Longhese MP. Marsella A, et al. Curr Genet. 2019 Feb;65(1):11-16. doi: 10.1007/s00294-018-0861-5. Epub 2018 Jun 19. Curr Genet. 2019. PMID: 29922906 Review.
Cited by
- Epigenetic regulation of genomic integrity.
Deem AK, Li X, Tyler JK. Deem AK, et al. Chromosoma. 2012 Apr;121(2):131-51. doi: 10.1007/s00412-011-0358-1. Epub 2012 Jan 17. Chromosoma. 2012. PMID: 22249206 Free PMC article. Review. - Do chromatin changes around a nascent double strand DNA break spread spherically into linearly non-adjacent chromatin?
Savic V. Savic V. Front Genet. 2013 Jul 19;4:139. doi: 10.3389/fgene.2013.00139. eCollection 2013. Front Genet. 2013. PMID: 23882282 Free PMC article. - Chromatin remodeling at DNA double-strand breaks.
Price BD, D'Andrea AD. Price BD, et al. Cell. 2013 Mar 14;152(6):1344-54. doi: 10.1016/j.cell.2013.02.011. Cell. 2013. PMID: 23498941 Free PMC article. Review. - Fusing telomeres with RNF8.
Jacobs JJ. Jacobs JJ. Nucleus. 2012 Mar 1;3(2):143-9. doi: 10.4161/nucl.19322. Epub 2012 Mar 1. Nucleus. 2012. PMID: 22555600 Free PMC article. Review. - Saccharomyces cerevisiae: a versatile eukaryotic system in virology.
Galao RP, Scheller N, Alves-Rodrigues I, Breinig T, Meyerhans A, Díez J. Galao RP, et al. Microb Cell Fact. 2007 Oct 10;6:32. doi: 10.1186/1475-2859-6-32. Microb Cell Fact. 2007. PMID: 17927824 Free PMC article.
References
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. - PubMed
- Arkady C, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 2003;5:675–679. - PubMed
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. - PubMed
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials