A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes - PubMed (original) (raw)
A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes
Daniel H Haft et al. PLoS Comput Biol. 2005 Nov.
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPRs) are a family of DNA direct repeats found in many prokaryotic genomes. Repeats of 21-37 bp typically show weak dyad symmetry and are separated by regularly sized, nonrepetitive spacer sequences. Four CRISPR-associated (Cas) protein families, designated Cas1 to Cas4, are strictly associated with CRISPR elements and always occur near a repeat cluster. Some spacers originate from mobile genetic elements and are thought to confer "immunity" against the elements that harbor these sequences. In the present study, we have systematically investigated uncharacterized proteins encoded in the vicinity of these CRISPRs and found many additional protein families that are strictly associated with CRISPR loci across multiple prokaryotic species. Multiple sequence alignments and hidden Markov models have been built for 45 Cas protein families. These models identify family members with high sensitivity and selectivity and classify key regulators of development, DevR and DevS, in Myxococcus xanthus as Cas proteins. These identifications show that CRISPR/cas gene regions can be quite large, with up to 20 different, tandem-arranged cas genes next to a repeat cluster or filling the region between two repeat clusters. Distinctive subsets of the collection of Cas proteins recur in phylogenetically distant species and correlate with characteristic repeat periodicity. The analyses presented here support initial proposals of mobility of these units, along with the likelihood that loci of different subtypes interact with one another as well as with host cell defensive, replicative, and regulatory systems. It is evident from this analysis that CRISPR/cas loci are larger, more complex, and more heterogeneous than previously appreciated.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
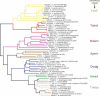
Figure 1. Distribution of the Different CRISPR/Cas Subtypes across Some of the Prokaryotic Species for Which a Whole-Genome Sequence Is Available
The taxonomy of each species/strain is indicated on the left side of the figure. The CRISPR/cas loci of a number of illustrative examples for the different CRISPR subtypes are displayed on the right side of the figure. a E. coli K12-MG1655, O157:H7 EDL933, and O157:H7 VT2-Sakai. b Salmonella enterica Paratyphi ATCC9150, serovar Typhi CT18, and Ty2; Salmonella typhimurium LT2 SGSC1412. c Y. pestis CO92, KIM, and biovar Mediaevalis 91001; Yersinia pseudotuberculosis IP32593. d“_p_” indicates a partial cluster lacking some of the genes usually associated with this subtype, the repeats, or both. Such clusters may represent autonomous functional units, degradation from the common subtype, or cases in which the missing components are supplied by distantly located CRISPR clusters within the same genome.
Figure 2. Molecular Phylogeny of the Cas1 Protein across 54 Prokaryotic Genomes
A representative selection of Cas1 protein sequences were aligned using ClustalW, and columns with greater than 20% gaps were removed. A neighbor-joining tree was calculated in Belvu using the Storm and Sonnhammer distance correction. Trees calculated using more computationally intensive methods showed insignificant differences. aFrom the preliminary annotation of the Haloferax volcanii genome, currently sequenced at The Institute for Genomic Research (
http://www.tigr.org/tdb/mdb/mdbinprogress.html
).
Similar articles
- Identification of genes that are associated with DNA repeats in prokaryotes.
Jansen R, Embden JD, Gaastra W, Schouls LM. Jansen R, et al. Mol Microbiol. 2002 Mar;43(6):1565-75. doi: 10.1046/j.1365-2958.2002.02839.x. Mol Microbiol. 2002. PMID: 11952905 - Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin.
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Bolotin A, et al. Microbiology (Reading). 2005 Aug;151(Pt 8):2551-2561. doi: 10.1099/mic.0.28048-0. Microbiology (Reading). 2005. PMID: 16079334 - The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes.
Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. Shmakov SA, et al. mBio. 2017 Sep 19;8(5):e01397-17. doi: 10.1128/mBio.01397-17. mBio. 2017. PMID: 28928211 Free PMC article. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - The CRISPR conundrum: evolve and maybe die, or survive and risk stagnation.
García-Martínez J, Maldonado RD, Guzmán NM, Mojica FJM. García-Martínez J, et al. Microb Cell. 2018 May 16;5(6):262-268. doi: 10.15698/mic2018.06.634. Microb Cell. 2018. PMID: 29850463 Free PMC article. Review.
Cited by
- CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus.
Manica A, Schleper C. Manica A, et al. RNA Biol. 2013 May;10(5):671-8. doi: 10.4161/rna.24154. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535277 Free PMC article. Review. - Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-Associated Protein and Its Utility All at Sea: Status, Challenges, and Prospects.
Li J, Wu S, Zhang K, Sun X, Lin W, Wang C, Lin S. Li J, et al. Microorganisms. 2024 Jan 6;12(1):118. doi: 10.3390/microorganisms12010118. Microorganisms. 2024. PMID: 38257946 Free PMC article. Review. - Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells.
Chen Y, Liu J, Zhi S, Zheng Q, Ma W, Huang J, Liu Y, Liu D, Liang P, Songyang Z. Chen Y, et al. Nat Commun. 2020 Jun 19;11(1):3136. doi: 10.1038/s41467-020-16880-8. Nat Commun. 2020. PMID: 32561716 Free PMC article. - Molecular identification and characterization of clustered regularly interspaced short palindromic repeat (CRISPR) gene cluster in Taylorella equigenitalis.
Hara Y, Hayashi K, Nakajima T, Kagawa S, Tazumi A, Moore JE, Matsuda M. Hara Y, et al. Folia Microbiol (Praha). 2013 Sep;58(5):375-84. doi: 10.1007/s12223-012-0217-3. Epub 2012 Dec 30. Folia Microbiol (Praha). 2013. PMID: 23275249 - Comparative Genomics Reveals Ecological and Evolutionary Insights into Sponge-Associated Thaumarchaeota.
Zhang S, Song W, Wemheuer B, Reveillaud J, Webster N, Thomas T. Zhang S, et al. mSystems. 2019 Aug 13;4(4):e00288-19. doi: 10.1128/mSystems.00288-19. mSystems. 2019. PMID: 31409660 Free PMC article.
References
- Mojica FJ, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. - PubMed
- Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. - PubMed
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. - PubMed
- Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. OMICS. 2002;6:23–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases