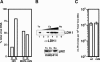The cell cycle independence of HIV infections is not determined by known karyophilic viral elements - PubMed (original) (raw)
The cell cycle independence of HIV infections is not determined by known karyophilic viral elements
Masahiro Yamashita et al. PLoS Pathog. 2005 Nov.
Abstract
Human immunodeficiency virus and other lentiviruses infect cells independent of cell cycle progression, but gammaretroviruses, such as the murine leukemia virus (MLV) require passage of cells through mitosis. This property is thought to be important for the ability of HIV to infect resting CD4+ T cells and terminally differentiated macrophages. Multiple and independent redundant nuclear localization signals encoded by HIV have been hypothesized to facilitate migration of viral genomes into the nucleus. The integrase (IN) protein of HIV is one of the HIV elements that targets to the nucleus; however, its role in nuclear entry of virus genomes has been difficult to describe because mutations in IN are pleiotropic. To investigate the importance of the HIV IN protein for infection of non-dividing cells, and to investigate whether or not IN was redundant with other viral signals for cell cycle-independent nuclear entry, we constructed an HIV-based chimeric virus in which the entire IN protein of HIV was replaced by that of MLV. This chimeric virus with a heterologous IN was infectious at a low level, and was able to integrate in an IN-dependent manner. Furthermore, this virus infected non-dividing cells as well as it infected dividing cells. Moreover, we used the chimeric HIV with MLV IN to further eliminate all of the other described nuclear localization signals from an HIV genome--matrix, IN, Viral Protein R, and the central polypurine tract--and show that no combination of the virally encoded NLS is essential for the ability of HIV to infect non-dividing cells.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Schematic Representation of the Genomic Organization of Chimeric HIV/MLV Proviruses
Portions originated from the HIV genome are shown in white, while those from the MLV genome are in gray. The junction between HIV-1 RT and MLV IN within HIV-mIN was created by direct joining of DNA sequences encoding the C-terminus of HIV RT to the N-terminus of MLV IN. Part of the 3′ end of the HIV-1 IN encoding sequence is retained in the construct of MHIV-mIN to preserve the overlapping Vif sequence and cis_-acting elements such as cPPT and splice acceptor(s). However, no part of HIV IN should be expressed in the chimeric virus because of the presence of two stop codons following the sequence encoding MLV IN (see Materials and Methods). The molecular clone encoding MHIV-mIN/m_att has the MLV att sites in 5′ U5 and 3′ U3. After reverse transcription, both ends of U5 and U3 will have the MLV att sites. MHIV-mMA12CA has been previously described [40]. MHIV-mMA12CA/mIN is similar except it contains MLV IN in addition to the MLV Gag region. A molecular clone of MHIV-mMA12CA/mIN was created by putting the DNA sequence encompassing the MLV IN encoding sequence of the MHIV-mIN with a Vpr mutation into the infectious provirus pMHIV-mMA12CA. HIVΔNLS contains MLV MA instead of HIV MA, MLV IN instead of HIV IN and mutations in the cPPT and in Vpr.
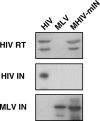
Figure 2. Western Blot Analysis of Purified Virus Particles of MHIV-mIN Together with HIV-1 and MLV
Polyprotein processing was tested for HIV-1 RT, HIV-1 IN and MLV IN. Because of low protein production by MHIV-mIN, 10-times more virions were loaded for MHIV-mIN than for HIV-1 and MLV in this experiment.
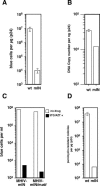
Figure 3. MHIV-mIN Is Infectious
(A) Single-cycle infectivity of MHIV-mIN. VSV-G-pseudotyped HIV and MHIV-mIN were made by transfection of 293T cells with plasmid DNA. Infectivity was measured with the MAGI assay by counting β-galactosidase positive cells 2 d post-infection. Virus titers were normalized by the amount of p24 (μg). Infections were performed in triplicate. Mean values are shown here with standard derivation. The background in the assay is about 10 blue cells. (B) Copy numbers of late products of reverse transcription were measured by using real-time quantitative PCR. Viral cDNA numbers were normalized by p24. Infections were performed in triplicate. This is a representative experiment of three independent trials. (C) Infectivity of MHIV-mIN and MHIV-mIN/m_att_ was compared in the MAGI assay. VSV-G-pseudotyped viruses were prepared and concentrated at 100-fold by ultracentrifugation. Infections of MAGI cells were also performed in the presence or absence of reverse transcriptase inhibitors (50 μM 3TC and AZT). Formation of blue cells in this assay must result from retrovirus infection since addition of reverse transcription inhibitors (shown as black bars) eliminated most of positive cells. (D) Comparison of infectivity between wild-type HIV-1 and MHIV-mIN as judged by the ability to make puromycin-resistant clones. Infectivity was measured by counting puromycin-resistant colonies 14 d after infection. To facilitate stable transduction of HIV genomes, a mutation was introduced into the vpr gene in these proviruses because expression of Vpr would preclude formation of colonies due to its cytotoxicity [69,77]
Figure 4. Integration Sites of MHIV-mIN
A schematic illustration of the unintegrated viral DNA is shown with the detailed structures of both ends of LTR (top). The two terminal base pairs at each end of the linear DNA precursor, which are removed in the integration process, as shown in highlight. Two other DNA sequences confirmed the removal of dinucleotides from one end of viral DNA (unpublished data). DNA sequences flanking integrated proviral DNA are shown (bottom). Junction sequences between the integrated MHIV-mIN genome and human genomic DNA at each end of the provirus were obtained by nested PCR based on the sequence of integration sites that were mapped with human genome sequences as described in the Materials and Methods. The 4-bp sequence duplications that flank the integrated provirus are shown in boxes.
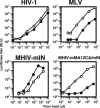
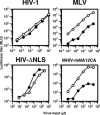
Figure 5. IN Does Not Determine the Infectivity in Non-Dividing Cells
Aphidicolin-treated HeLa cells were infected with increasing amount of luciferase-encoding viruses described in Figure 1. Culture supernatants of transfected cells were used as the inoculum. Virus infectivity was judged by measuring luciferase titers of infected cell lysates 2 d after infection. RLU; relative light units. White circles indicate cells without aphidicolin. Filled circles indicate cells with aphidicolin (2 μg per ml). This is a representative experiment that was done at least three times for each virus.
Figure 6. Nuclear Entry of MHIV-mIN
(A) Subcellular localization of viral DNA. Infected cells were fractionated to cytoplasmic (C) and nuclear (N) fractions. Viral DNA was extracted and subject to real-time PCR to measure late products of reverse transcription. These data represent one of two independent experiments. Control viruses, with the presence of reverse transcription inhibitors or without VSV-G protein, were also used in the experiments to monitor retrovirus-dependent DNA synthesis, and showed that contamination of plasmid DNA used for transfection is less than 1% of the total DNA. (B) Western blot analysis of total cell lysates (To), cytoplasmic extract (Cy) and nuclear lysates (Nu). Contamination of cytoplasmic extract and the presence of intact cells in nuclear fractions were tested by checking for the presence of a cytoplasmic protein, LDH-I, in each fraction (upper lanes). Five-fold dilutions of the cytoplasmic extract (5-, 25-, and 125-fold dilutions, lanes 4, 3, and 2, respectively) were made to assess the degree of contamination of the nuclear fraction. Presence of proteins was confirmed by antibody against a nuclear pore complex protein (mAb414; lower lane). (C) Nuclear import was monitored by measuring late reverse transcription products and 2-LTR circles. The ratio of total viral DNA and 2-LTR circles was obtained by dividing the copy number of late RT products by the copy number of 2-LTR circle. The parental wild-type strain of HIV-1 (shown here as wt) was compared with the chimeric virus MHIV-mIN (shown as mIN). Control infections with reverse transcription inhibitors (AZT and 3TC: 50 μM each) yielded viral copy numbers that are less than 10% of copy numbers of the samples without reverse transcription inhibitors, indicating that contamination of plasmid DNA used to produce virus stocks does not affect the final results. Two independent experiments gave substantially identical data.
Figure 7. HIV Lacking NLSs Infects Non-Dividing Cells
Single-cycle infectivity assay of HIV-ΔNLS together with HIV, MLV, and MHIV-mMA12CA. Both HIV-ΔNLS and MHIV-mMA12CA were concentrated by ultracentrifugation for infections. For details, see the legend to Figure 5. White circles indicate cells without aphidicolin. Filled circles indicate cells with aphidicolin (2 μg per ml). This is a representative experiment that was done at least 3 times for each virus.
Similar articles
- Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses.
Depienne C, Roques P, Créminon C, Fritsch L, Casseron R, Dormont D, Dargemont C, Benichou S. Depienne C, et al. Exp Cell Res. 2000 Nov 1;260(2):387-95. doi: 10.1006/excr.2000.5016. Exp Cell Res. 2000. PMID: 11035935 - Nuclear localization of Vpr is crucial for the efficient replication of HIV-1 in primary CD4+ T cells.
Iijima S, Nitahara-Kasahara Y, Kimata K, Zhong Zhuang W, Kamata M, Isogai M, Miwa M, Tsunetsugu-Yokota Y, Aida Y. Iijima S, et al. Virology. 2004 Oct 1;327(2):249-61. doi: 10.1016/j.virol.2004.06.024. Virology. 2004. PMID: 15351213 - Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex.
Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, Nadler SG, Bukrinsky M. Haffar OK, et al. J Mol Biol. 2000 Jun 2;299(2):359-68. doi: 10.1006/jmbi.2000.3768. J Mol Biol. 2000. PMID: 10860744 - Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV?
Piller SC, Caly L, Jans DA. Piller SC, et al. Curr Drug Targets. 2003 Jul;4(5):409-29. doi: 10.2174/1389450033490984. Curr Drug Targets. 2003. PMID: 12816349 Review. - Insights into the biology of HIV-1 viral protein R.
Sherman MP, De Noronha CM, Williams SA, Greene WC. Sherman MP, et al. DNA Cell Biol. 2002 Sep;21(9):679-88. doi: 10.1089/104454902760330228. DNA Cell Biol. 2002. PMID: 12396611 Review.
Cited by
- The importance of becoming double-stranded: Innate immunity and the kinetic model of HIV-1 central plus strand synthesis.
Poeschla E. Poeschla E. Virology. 2013 Jun 20;441(1):1-11. doi: 10.1016/j.virol.2013.03.010. Epub 2013 Apr 3. Virology. 2013. PMID: 23561461 Free PMC article. Review. - Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74.
Saito A, Ferhadian D, Sowd GA, Serrao E, Shi J, Halambage UD, Teng S, Soto J, Siddiqui MA, Engelman AN, Aiken C, Yamashita M. Saito A, et al. J Virol. 2016 May 27;90(12):5808-5823. doi: 10.1128/JVI.03116-15. Print 2016 Jun 15. J Virol. 2016. PMID: 27076642 Free PMC article. - Effects of human T-cell leukemia virus type 1 (HTLV-1) p13 on mitochondrial K+ permeability: A new member of the viroporin family?
Silic-Benussi M, Marin O, Biasiotto R, D'Agostino DM, Ciminale V. Silic-Benussi M, et al. FEBS Lett. 2010 May 17;584(10):2070-5. doi: 10.1016/j.febslet.2010.02.030. Epub 2010 Feb 17. FEBS Lett. 2010. PMID: 20170654 Free PMC article. Review. - Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells.
Yamashita M, Perez O, Hope TJ, Emerman M. Yamashita M, et al. PLoS Pathog. 2007 Oct 26;3(10):1502-10. doi: 10.1371/journal.ppat.0030156. PLoS Pathog. 2007. PMID: 17967060 Free PMC article. - A cyclophilin homology domain-independent role for Nup358 in HIV-1 infection.
Meehan AM, Saenz DT, Guevera R, Morrison JH, Peretz M, Fadel HJ, Hamada M, van Deursen J, Poeschla EM. Meehan AM, et al. PLoS Pathog. 2014 Feb 20;10(2):e1003969. doi: 10.1371/journal.ppat.1003969. eCollection 2014 Feb. PLoS Pathog. 2014. PMID: 24586169 Free PMC article.
References
- Desrosiers R. Nonhuman lentiviruses. 2001. In: Knipe D, Howley P, Griffin D, Lamb R, Martin M et al., editors. Fields Virology. 4 ed. Philadephia: Lippincott, Williams, and Wilkins. pp. 2095–2121.
- Stevenson M. HIV-1 pathogenesis. Nat Med. 2003;9:853–860. - PubMed
- Katz RA, Greger JG, Skalka AM. Effects of cell cycle status on early events in retroviral replication. J Cell Biochem. 2005;94:880–889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI051153/AI/NIAID NIH HHS/United States
- R37 AI030927/AI/NIAID NIH HHS/United States
- R01 AI 51153/AI/NIAID NIH HHS/United States
- R37 AI 30927/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials