An acquisition account of genomic islands based on genome signature comparisons - PubMed (original) (raw)
An acquisition account of genomic islands based on genome signature comparisons
M W J van Passel et al. BMC Genomics. 2005.
Abstract
Background: Recent analyses of prokaryotic genome sequences have demonstrated the important force horizontal gene transfer constitutes in genome evolution. Horizontally acquired sequences are detectable by, among others, their dinucleotide composition (genome signature) dissimilarity with the host genome. Genomic islands (GIs) comprise important and interesting horizontally transferred sequences, but information about acquisition events or relatedness between GIs is scarce. In Vibrio vulnificus CMCP6, 10 and 11 GIs have previously been identified in the sequenced chromosomes I and II, respectively. We assessed the compositional similarity and putative acquisition account of these GIs using the genome signature. For this analysis we developed a new algorithm, available as a web application.
Results: Of 21 GIs, VvI-1 and VvI-10 of chromosome I have similar genome signatures, and while artificially divided due to a linear annotation, they are adjacent on the circular chromosome and therefore comprise one GI. Similarly, GIs VvI-3 and VvI-4 of chromosome I together with the region between these two islands are compositionally similar, suggesting that they form one GI (making a total of 19 GIs in chromosome I + chromosome II). Cluster analysis assigned the 19 GIs to 11 different branches above our conservative threshold. This suggests a limited number of compositionally similar donors or intragenomic dispersion of ancestral acquisitions. Furthermore, 2 GIs of chromosome II cluster with chromosome I, while none of the 19 GIs group with chromosome II, suggesting an unidirectional dispersal of large anomalous gene clusters from chromosome I to chromosome II.
Conclusion: From the results, we infer 10 compositionally dissimilar donors for 19 GIs in the V. vulnificus CMCP6 genome, including chromosome I donating to chromosome II. This suggests multiple transfer events from individual donor types or from donors with similar genome signatures. Applied to other prokaryotes, this approach may elucidate the acquisition account in their genome sequences, and facilitate donor identification of GIs.
Figures
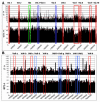
Figure 1
Overview of the two V. vulnificus chromosomes. Schematic representation of the δ* values and GC content of large putative horizontally transferred gene clusters in A) chromosome I and B) chromosome II of V. vulnificus CMCP6 using a window size of 5 kbp (x-axis represents chromosome position). Red depicts the low GC content GIs, while blue depicts the high GC content GIs. In green, a large ribosomal protein gene cluster is depicted (Rib). The horizontal dashed red line represents the average δ* value and GC percentage, respectively.
Figure 2
Hierarchic clustering with complete linkage of the V. vulnificus GIs (as described in table 1) based on the genome signature. Three non-anomalous genomic fragments (indicated with Vv5%, Vv10% and Vv25%) represent the conservative V. vulnificus (VvI) genomic variability, and this clade forms the cut-off value for the different clades (with the red dotted line; clades are indicated with black boxes). The Chlamydia clade consists of two genomic fragments (Ct1 and Ct2) and the genome sequence of C. trachomatis. VvII represents V. vulnificus chromosome II.
Figure 3
Hierarchic clustering with complete linkage of the V. vulnificus GIs from both chromosomes (as described in tables 1 and 2) based on the genome signature. For both chromosomes three non-anomalous genomic fragments are included, which represent the conservative V. vulnificus (VvI and VvII) genomic variability. VvI and VvII represent V. vulnificus chromosome I and II, respectively. VvI-3inter4 and VvI-101 represent the concatenated islands of VvI-3, VvI-inter and VvI-4 and of VvI10 and VvI-1 respectively.
Similar articles
- Genomic island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen.
Quirke AM, Reen FJ, Claesson MJ, Boyd EF. Quirke AM, et al. Bioinformatics. 2006 Apr 15;22(8):905-10. doi: 10.1093/bioinformatics/btl015. Epub 2006 Jan 27. Bioinformatics. 2006. PMID: 16443635 - A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I.
Zhang R, Zhang CT. Zhang R, et al. Bioinformatics. 2004 Mar 22;20(5):612-22. doi: 10.1093/bioinformatics/btg453. Epub 2004 Jan 22. Bioinformatics. 2004. PMID: 15033867 - Involvement of β-Carbonic Anhydrase Genes in Bacterial Genomic Islands and Their Horizontal Transfer to Protists.
Zolfaghari Emameh R, Barker HR, Hytönen VP, Parkkila S. Zolfaghari Emameh R, et al. Appl Environ Microbiol. 2018 Jul 17;84(15):e00771-18. doi: 10.1128/AEM.00771-18. Print 2018 Aug 1. Appl Environ Microbiol. 2018. PMID: 29802189 Free PMC article. - A quantitative account of genomic island acquisitions in prokaryotes.
Roos TE, van Passel MW. Roos TE, et al. BMC Genomics. 2011 Aug 24;12:427. doi: 10.1186/1471-2164-12-427. BMC Genomics. 2011. PMID: 21864345 Free PMC article. - Detecting genomic islands using bioinformatics approaches.
Langille MG, Hsiao WW, Brinkman FS. Langille MG, et al. Nat Rev Microbiol. 2010 May;8(5):373-82. doi: 10.1038/nrmicro2350. Nat Rev Microbiol. 2010. PMID: 20395967 Review.
Cited by
- Genomic Signature in Evolutionary Biology: A Review.
de la Fuente R, Díaz-Villanueva W, Arnau V, Moya A. de la Fuente R, et al. Biology (Basel). 2023 Feb 16;12(2):322. doi: 10.3390/biology12020322. Biology (Basel). 2023. PMID: 36829597 Free PMC article. Review. - A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes.
Cortez D, Forterre P, Gribaldo S. Cortez D, et al. Genome Biol. 2009;10(6):R65. doi: 10.1186/gb-2009-10-6-r65. Epub 2009 Jun 16. Genome Biol. 2009. PMID: 19531232 Free PMC article. - The use of genomic signature distance between bacteriophages and their hosts displays evolutionary relationships and phage growth cycle determination.
Deschavanne P, DuBow MS, Regeard C. Deschavanne P, et al. Virol J. 2010 Jul 17;7:163. doi: 10.1186/1743-422X-7-163. Virol J. 2010. PMID: 20637121 Free PMC article. - The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes.
Bohlin J, Eldholm V, Pettersson JH, Brynildsrud O, Snipen L. Bohlin J, et al. BMC Genomics. 2017 Feb 10;18(1):151. doi: 10.1186/s12864-017-3543-7. BMC Genomics. 2017. PMID: 28187704 Free PMC article. - Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus.
Mallet LV, Becq J, Deschavanne P. Mallet LV, et al. BMC Genomics. 2010 Mar 12;11:171. doi: 10.1186/1471-2164-11-171. BMC Genomics. 2010. PMID: 20226043 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources