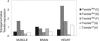Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice - PubMed (original) (raw)
Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice
Henna Tyynismaa et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Defects of mitochondrial DNA (mtDNA) maintenance have recently been associated with inherited neurodegenerative and muscle diseases and the aging process. Twinkle is a nuclear-encoded mtDNA helicase, dominant mutations of which cause adult-onset progressive external ophthalmoplegia (PEO) with multiple mtDNA deletions. We have generated transgenic mice expressing mouse Twinkle with PEO patient mutations. Multiple mtDNA deletions accumulate in the tissues of these mice, resulting in progressive respiratory dysfunction and chronic late-onset mitochondrial disease starting at 1 year of age. The muscles of the mice faithfully replicate all of the key histological, genetic, and biochemical features of PEO patients. Furthermore, the mice have progressive deficiency of cytochrome c oxidase in distinct neuronal populations. These "deletor" mice do not, however, show premature aging, indicating that subtle accumulation of mtDNA deletions and progressive respiratory chain dysfunction are not sufficient to accelerate aging. This model is a valuable tool for therapy development and testing for adult-onset mitochondrial disorders.
Figures
Fig. 1.
Transgene expression levels in the muscle, heart, and brain of the three transgenic Twinkledup (C, D, and F) and two TwinkleAT (G, H) mouse lines. The transgene expression levels were correlated with the expression of the native Twinkle gene.
Fig. 2.
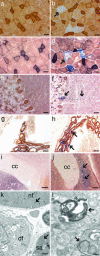
Cytochrome c oxidase activity of muscle and brain and the skeletal muscle ultrastructure. Double staining for COX/SDH activities of the quadriceps femoris of 12-month-old control (a) and Twinkledup mouse (b), the quadriceps femoris of 18-month-old control (c) and Twinkledup (d) mouse, cerebellum of 18-month-old control (e) and Twinkledup (f) mouse, choroid plexus of 18-month-old control (g) and Twinkledup (h) mouse, and indusium griseum of 18-month-old control (i) and Twinkledup (j) mouse (cc, corpus callosum). The arrows point to the affected cells of the transgenic mice. (k and l) Electron micrographs of ultrathin section of the quadriceps femoris of an 18-month-old Twinkledup mouse. (k) Normal-sized mitochondrion is pointed out by the arrow in the normal fiber (nf) and an enlarged mitochondrion in the fiber with subsarcolemmal accumulation of mitochondria (sa). In the diseased fiber (df), muscle fibrils have been replaced by abnormal large mitochondria with inclusions, peripheral cristae, and vacuoles. (l) The autophagosomes with mitochondria found from the diseased fibers are shown by arrows. (Scale bars: 25 μm, a–h; 100 μm, i and j; 1 μm, k; 200 nm, l.)
Fig. 3.
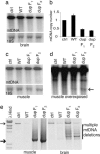
Mitochondrial DNA analysis and copy number determination. (a and c) Southern blots of muscle and brain DNA of 22-month-old control, TwinkleWT+ mice, and Twinkledup F1 mouse and an 18-month-old Twinkledup F2 mice probed with mtDNA, and nuclear 18S rRNA gene. (b) Quantitation of brain mtDNA copy number of the mice against the nuclear 18S rRNA gene. (d) Overexposed Southern blot of muscle DNA shown in c. The 3-kb band found from Twinkledup mice is pointed by the arrow. (e) Long PCR of muscle and brain DNA of the same mice. The full-length mtDNA is indicated by the thin arrow and the most prevalent deletion molecule size of muscle mtDNA (≈3 kb) by the thick arrow.
Fig. 4.
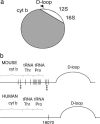
mtDNA deletions of the muscle and brain of the Twinkledup mouse, determined by long PCR, cloning, and DNA sequencing. (a) Schematic diagram of mouse mtDNA showing the deleted segment in gray (see
supporting information
for details). Only the D-loop, 12S rRNA, and 16S rRNA were commonly spared. (b) Schematic presentation of the mouse and human mtDNA region harboring the 5′ deletion breakpoints. Black vertical lines show the 5′ deletion breakpoints identified in this study, and the asterisks below indicate how often the same 5′ breakpoint was found from dissimilar deletion molecules. The common 5′deletion breakpoint of human multiple mtDNA deletions (16070) is indicated.
Similar articles
- Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling.
Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Goffart S, et al. Hum Mol Genet. 2009 Jan 15;18(2):328-40. doi: 10.1093/hmg/ddn359. Epub 2008 Oct 29. Hum Mol Genet. 2009. PMID: 18971204 Free PMC article. - Mitochondrial myopathy induces a starvation-like response.
Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkilä S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjøsund KE, Tulkki V, Oresic M, Moraes CT, Pietiläinen K, Hovatta I, Suomalainen A. Tyynismaa H, et al. Hum Mol Genet. 2010 Oct 15;19(20):3948-58. doi: 10.1093/hmg/ddq310. Epub 2010 Jul 23. Hum Mol Genet. 2010. PMID: 20656789 - POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions.
Di Fonzo A, Bordoni A, Crimi M, Sara G, Del Bo R, Bresolin N, Comi GP. Di Fonzo A, et al. Hum Mutat. 2003 Dec;22(6):498-9. doi: 10.1002/humu.9203. Hum Mutat. 2003. PMID: 14635118 - Progressive external ophthalmoplegia and multiple mitochondrial DNA deletions.
Van Goethem G, Martin JJ, Van Broeckhoven C. Van Goethem G, et al. Acta Neurol Belg. 2002 Mar;102(1):39-42. Acta Neurol Belg. 2002. PMID: 12094562 Review. - Finding twinkle in the eyes of a 71-year-old lady: a case report and review of the genotypic and phenotypic spectrum of TWINKLE-related dominant disease.
Van Hove JL, Cunningham V, Rice C, Ringel SP, Zhang Q, Chou PC, Truong CK, Wong LJ. Van Hove JL, et al. Am J Med Genet A. 2009 May;149A(5):861-7. doi: 10.1002/ajmg.a.32731. Am J Med Genet A. 2009. PMID: 19353676 Review.
Cited by
- Mitochondrial Diseases Part III: Therapeutic interventions in mouse models of OXPHOS deficiencies.
Peralta S, Torraco A, Iommarini L, Diaz F. Peralta S, et al. Mitochondrion. 2015 Jul;23:71-80. doi: 10.1016/j.mito.2015.01.007. Epub 2015 Jan 29. Mitochondrion. 2015. PMID: 25638392 Free PMC article. Review. - An atypical case of neuronal ceroid lipofuscinosis with co-inheritance of a variably penetrant POLG1 mutation.
Staropoli JF, Xin W, Barone R, Cotman SL, Sims KB. Staropoli JF, et al. BMC Med Genet. 2012 Jun 24;13:50. doi: 10.1186/1471-2350-13-50. BMC Med Genet. 2012. PMID: 22727047 Free PMC article. - The role of mitochondrial DNA mutations and free radicals in disease and ageing.
Lagouge M, Larsson NG. Lagouge M, et al. J Intern Med. 2013 Jun;273(6):529-43. doi: 10.1111/joim.12055. Epub 2013 Mar 7. J Intern Med. 2013. PMID: 23432181 Free PMC article. Review. - The different axes of the mammalian mitochondrial unfolded protein response.
Münch C. Münch C. BMC Biol. 2018 Jul 26;16(1):81. doi: 10.1186/s12915-018-0548-x. BMC Biol. 2018. PMID: 30049264 Free PMC article. Review. - Helicase-inactivating mutations as a basis for dominant negative phenotypes.
Wu Y, Brosh RM Jr. Wu Y, et al. Cell Cycle. 2010 Oct 15;9(20):4080-90. doi: 10.4161/cc.9.20.13667. Cell Cycle. 2010. PMID: 20980836 Free PMC article. Review.
References
- Zeviani, M., Servidei, S., Gellera, C., Bertini, E., DiMauro, S. & DiDonato, S. (1989) Nature 339, 309–311. - PubMed
- Van Goethem, G., Dermaut, B., Lofgren, A., Martin, J. J. & Van Broeckhoven, C. (2001) Nat. Genet. 28, 211–212. - PubMed
- Van Goethem, G., Luoma, P., Rantamaki, M., Al Memar, A., Kaakkola, S., Hackman, P., Krahe, R., Lofgren, A., Martin, J. J., De Jonghe, P., et al. (2004) Neurology 63, 1251–1257. - PubMed
- Luoma, P., Melberg, A., Rinne, J. O., Kaukonen, J. A., Nupponen, N. N., Chalmers, R. M., Oldfors, A., Rautakorpi, I., Peltonen, L., Majamaa, K. et al. (2004) Lancet 364, 875–882. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases