Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1 - PubMed (original) (raw)
Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1
Inmaculada Hernández-Muñoz et al. Mol Cell Biol. 2005 Dec.
Abstract
Polycomb group (PcG) proteins are epigenetic chromatin modifiers involved in heritable gene repression. Two main PcG complexes have been characterized. Polycomb repressive complex 2 (PRC2) is thought to be involved in the initiation of gene silencing, whereas Polycomb repressive complex 1 (PRC1) is implicated in the stable maintenance of gene repression. Here, we investigate the kinetic properties of the binding of one of the PRC1 core components, BMI1, with PcG bodies. PcG bodies are unique nuclear structures located on regions of pericentric heterochromatin, found to be the site of accumulation of PcG complexes in different cell lines. We report the presence of at least two kinetically different pools of BMI1, a highly dynamic and a less dynamic fraction, which may reflect BMI1 pools with different binding capacities to these stable heterochromatin domains. Interestingly, PRC2 members EED and EZH2 appear to be essential for BMI1 recruitment to the PcG bodies. Furthermore, we demonstrate that the maintenance DNA methyltransferase DNMT1 is necessary for proper PcG body assembly independent of DNMT-associated histone deacetylase activity. Together, these results provide new insights in the mechanism for regulation of chromatin silencing by PcG proteins and suggest a highly regulated recruitment of PRC1 to chromatin.
Figures
FIG. 1.
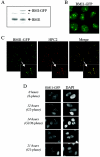
GFP-tagged BMI1 is correctly targeted to PcG bodies and closely resembles the endogenous BMI1 localization. (A) BMI1-GFP expression level is close to endogenous BMI1 expression level. Western blot analysis was conducted using extracts from parental and BMI1-GFP-expressing U2OS cells, hybridized with the BMI1 antibody. *, nonspecific band. (B) BMI1-GFP protein is targeted to PcG bodies, PcG-related pericentric heterochromatin foci, in U2OS cells as shown by GFP fluorescence. In addition, lower levels of green fluorescence in the regions between these foci are detected, indicating that BMI1-GFP is also present in euchromatic areas. (C) Confocal fluorescence microscopy demonstrating colocalization of BMI1-GFP with HPC2 (polyclonal anti-HPC2 immunostaining) in large nuclear domains (PcG bodies) of U2OS cells stably expressing BMI1-GFP. (D) Cell cycle-dependent chromatin association of BMI1-GFP in U2OS cells closely resembling endogenous BMI1 association (57). Cells were synchronized in early S phase by a double thymidine block and subsequently released. Representative fluorescence images were made at indicated time points after release.
FIG. 2.
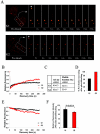
BMI1 is dynamically associated with PcG bodies. (A) Representative examples of a FRAP profile from BMI1-GFP-expressing U2OS cells. Cells were synchronized in G1 by a nocodazole treatment followed by release from the blockade or in G2 by a double thymidine block and subsequent release. Pseudo-colored images show the fluorescence signals and recovery with time. Arrowheads point to the bleached PcG bodies. (B) FRAP plots. Recovery of fluorescence in the bleached spots was plotted with time for the G1 and G2 cell cycle phases. The prebleach fluorescence was set at 100%, and the fluorescence intensity for each time point was averaged and normalized relative to the prebleach intensities after correcting for background and bleaching during the scanning procedure. (C) Quantifications of the recovery time (_t_1/2) and mobile fraction deduced from the best-fit analysis of the recovery plots. BMI1 has a slightly reduced _t_1/2 and mobile fraction in G1 versus G2 cell cycle phase. (D) Percentage of PcG bodies that showed any BMI1-GFP fluorescence recovery after photobleaching, indicating that BMI1-GFP is considerably less mobile in G1 than in G2. (E) FLIP plots. Loss of fluorescence with time from PcG bodies in a nonbleached region after photobleaching of a defined area of the nucleus was plotted for the G1 and G2 cell cycle phases. The prebleach fluorescence was set at 100%, and the fluorescence intensity for each time point was averaged and normalized relative to the prebleach intensities after correcting for background and bleaching during the scanning procedure. (F) Relative left fluorescence intensities at 240 s postbleaching are shown for G1 and G2 (±standard error of the means); prebleach fluorescence intensity was set at 100%. Loss of fluorescence in G2 is significantly faster than in G1, indicating the presence of a more mobile BMI1-GFP fraction on PcG bodies in G2.
FIG. 3.
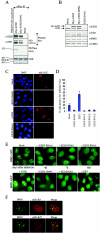
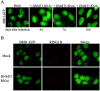
PRC2 member EZH2 is required for PcG body association of BMI1. (A) EZH2-H694L (SET domain mutant) is enzymatically inactive but still able to interact with EED. HEK293 cells were transiently transfected with EZH2 or EZH2-H694L or were mock transfected. Anti-Myc immunoprecipitated complexes were used for in vitro HMTase assays on core histone, shown in the third row from the top of the panel. The same immunoprecipitated complexes were probed with EZH2 and EED antibodies to confirm the pull-downs and functional PRC2 complex formation (the two upper rows). The bottom row shows Coomassie brilliant blue (CBB) staining for equal loading of the HMTase assay. (B) Western blot analysis showing expression levels of EZH2, EED, and BMI1 in U2OS cells used for immunostainings. Anti-CDK4 is used to show equal loading. Note that EZH2-H694L is moderately overexpressed compared to endogenous EZH2. (C) EZH2-H694L works in a dominant-interfering manner. Representative immunofluorescence images of U2OS cells transduced with EZH2, EZH2-H694L, or EZH2 RNAi1 or mock treated, using anti-trimethyl-H3-K27 (mH3-K27), indicating that EZH2-H694L is able to reduce the H3-K27 methylation levels in vivo by competing with endogenous wild-type EZH2. (D) Graph depicting quantifications of mH3-K27 immunofluorescence in U2OS cells transduced with EZH2, EZH2-H694L, or EZH2 RNAi1-3 or mock treated. Cells with mH3-K27foci were counted (n > 500 cells counted per condition from three independent experiments ± standard deviation). (E) Epifluorescence images of U2OS cells stably expressing BMI1-GFP, after transduction with EZH2 RNAi or mock treatment for the times indicated (upper row) or with EZH2, EZH2-H694L, EED, and EED RNAi1 10 days after transduction and selection with puromycin (lower row). (F) Immunofluorescence images of U2OS cells stained for di- and trimethylated H3-K9 or trimethylated H3-K27 and costained with anti-BMI1. Confocal single optical sections are shown. Note that PcG-associated heterochromatin is differentially recognized by antibodies against H3-K9 and H3-K27 methylation.
FIG. 4.
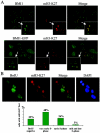
H3-K27 trimethylation colocalizes with Polycomb bodies in a cell cycle-dependent manner. (A) Asynchronously growing parental and BMI1-GFP-expressing U2OS cells were fixed and stained with mH3-K27 antibody and with the BMI1 antibody (upper row, parental U2OS cells). Merges of BMI1 and mH3-K27 signals show perfect overlap of the mH3-K27 with the PcG bodies. (B) Asynchronously growing U2OS cells were treated with 10 μM BrdU for 1 h. The cells were fixed and stained with antibodies against BrdU and mH3-K27 and with DAPI (see Materials and Methods). Based on the staining of the incorporated BrdU into the nascent DNA, different stages of S phase were recognized. In the figure representative images of different patterns of distribution of replication sites are shown. In early S phase, replication sites are distributed throughout the nucleus. As cells proceed from early to mid S phase, nuclei show no unlabeled areas and a fairly uniform staining of the nucleus. In the late S phase, replication of the bulk of heterochromatin follows, appearing as a pattern of large discrete foci. More than 200 cells positive for mH3-K27 foci were scored from at least eight random fields and classified into these different stages of the S phase.
FIG. 5.
Reduced DNMT1 levels disrupt BMI1 and RING1B recruitment to PcG body-associated pericentric heterochromatin domains. (A) U2OS cells stably expressing BMI1-GFP were transduced with DNMT1 RNAi and selected with puromycin. Cells were fixed at different time points (4, 7, or 10 days posttransduction), and the fluorescence was analyzed. (B) U2OS cells stably expressing BMI1-GFP were transduced with DNMT1 RNAi, and selected with puromycin. Cells were fixed 10 days posttransduction and stained with anti-RING1B to detect endogenous RING1B localization.
FIG. 6.
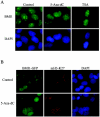
BMI1 localization to PcG bodies is disrupted by the DNA methylation inhibitor 5-Aza-dC, whereas H3-K27 methylation is not affected by DNA methylation inhibitor 5-Aza-dC. (A) Representative immunofluorescence images of parental U2OS cells treated with 5-Aza-dC or with TSA for 4 days, fixed, and stained with BMI1 antibody. BMI1 localization on PcG bodies is not affected by the HDAC inhibition. However, it is severely disrupted in cells treated with the DNA methylation inhibitor. (B) Untreated and 5-Aza-dC-treated BMI1-GFP-expressing U2OS cells were immunostained with mH3-K27 antibody. Representative immunofluorescence images show H3-K27 trimethylation persisting after 5-Aza-dC treatment. Whereas BMI1-GFP fluorescence is dispersed in the nucleus in the 5-Aza-dC-treated cells, the nuclear distribution of mH3-K27 is clearly not affected.
Similar articles
- The Polycomb group protein EZH2 directly controls DNA methylation.
Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. Viré E, et al. Nature. 2006 Feb 16;439(7078):871-4. doi: 10.1038/nature04431. Epub 2005 Dec 14. Nature. 2006. PMID: 16357870 - Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features.
van Leenders GJ, Dukers D, Hessels D, van den Kieboom SW, Hulsbergen CA, Witjes JA, Otte AP, Meijer CJ, Raaphorst FM. van Leenders GJ, et al. Eur Urol. 2007 Aug;52(2):455-63. doi: 10.1016/j.eururo.2006.11.020. Epub 2006 Nov 17. Eur Urol. 2007. PMID: 17134822 - A Noncanonical Function of Polycomb Repressive Complexes Promotes Human Cytomegalovirus Lytic DNA Replication and Serves as a Novel Cellular Target for Antiviral Intervention.
Svrlanska A, Reichel A, Schilling EM, Scherer M, Stamminger T, Reuter N. Svrlanska A, et al. J Virol. 2019 Apr 17;93(9):e02143-18. doi: 10.1128/JVI.02143-18. Print 2019 May 1. J Virol. 2019. PMID: 30814291 Free PMC article. - Epithelial-mesenchymal transition and cancer stemness: the Twist1-Bmi1 connection.
Wu KJ, Yang MH. Wu KJ, et al. Biosci Rep. 2011 Dec;31(6):449-55. doi: 10.1042/BSR20100114. Biosci Rep. 2011. PMID: 21919891 Review. - The Role of Polycomb Group Protein BMI1 in DNA Repair and Genomic Stability.
Fitieh A, Locke AJ, Motamedi M, Ismail IH. Fitieh A, et al. Int J Mol Sci. 2021 Mar 15;22(6):2976. doi: 10.3390/ijms22062976. Int J Mol Sci. 2021. PMID: 33804165 Free PMC article. Review.
Cited by
- Fine structure of the "PcG body" in human U-2 OS cells established by correlative light-electron microscopy.
Smigová J, Juda P, Cmarko D, Raška I. Smigová J, et al. Nucleus. 2011 May-Jun;2(3):219-28. doi: 10.4161/nucl.2.3.15737. Nucleus. 2011. PMID: 21818415 Free PMC article. - Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications.
van de Werken C, van der Heijden GW, Eleveld C, Teeuwssen M, Albert M, Baarends WM, Laven JS, Peters AH, Baart EB. van de Werken C, et al. Nat Commun. 2014 Dec 18;5:5868. doi: 10.1038/ncomms6868. Nat Commun. 2014. PMID: 25519718 Free PMC article. - Cellular senescence and tumor suppressor gene p16.
Rayess H, Wang MB, Srivatsan ES. Rayess H, et al. Int J Cancer. 2012 Apr 15;130(8):1715-25. doi: 10.1002/ijc.27316. Epub 2011 Dec 5. Int J Cancer. 2012. PMID: 22025288 Free PMC article. Review. - Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis.
Sharma S, Mandal P, Sadhukhan T, Roy Chowdhury R, Ranjan Mondal N, Chakravarty B, Chatterjee T, Roy S, Sengupta S. Sharma S, et al. Sci Rep. 2015 Jul 8;5:11724. doi: 10.1038/srep11724. Sci Rep. 2015. PMID: 26152361 Free PMC article. - Nuclear Vav3 is required for polycomb repression complex-1 activity in B-cell lymphoblastic leukemogenesis.
Nayak RC, Chang KH, Singh AK, Kotliar M, Desai M, Wellendorf AM, Wunderlich M, Bartram J, Mizukawa B, Cuadrado M, Dexheimer P, Barski A, Bustelo XR, Nassar NN, Cancelas JA. Nayak RC, et al. Nat Commun. 2022 Jun 1;13(1):3056. doi: 10.1038/s41467-022-30651-7. Nat Commun. 2022. PMID: 35650206 Free PMC article.
References
- Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. - PubMed
- Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. - PubMed
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
- Bruggeman, S. W., M. E. Valk-Lingbeek, P. P. van der Stoop, J. J. Jacobs, K. Kieboom, E. Tanger, D. Hulsman, C. Leung, Y. Arsenijevic, S. Marino, and M. van Lohuizen. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19:1438-1443. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources