The ins and outs of DNA transfer in bacteria - PubMed (original) (raw)
Review
The ins and outs of DNA transfer in bacteria
Inês Chen et al. Science. 2005.
Abstract
Transformation and conjugation permit the passage of DNA through the bacterial membranes and represent dominant modes for the transfer of genetic information between bacterial cells or between bacterial and eukaryotic cells. As such, they are responsible for the spread of fitness-enhancing traits, including antibiotic resistance. Both processes usually involve the recognition of double-stranded DNA, followed by the transfer of single strands. Elaborate molecular machines are responsible for negotiating the passage of macromolecular DNA through the layers of the cell surface. All or nearly all the machine components involved in transformation and conjugation have been identified, and here we present models for their roles in DNA transport.
Figures
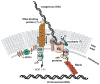
Fig. 1
Comparison of DNA processing and transfer during transformation and conjugation. (A) In transformation, dsDNA substrates are converted to single-stranded transfer intermediates for transport across the cytoplasmic membrane. (B) For conjugation, surface adhesins or conjugative pili mediate donor-target cell contacts. Initial reactions involve the formation of a relaxase–T-DNA transfer intermediate (green dot joined to black line) and tight mating junctions. Substrate transfer is probably mechanistically conserved in bacteria, although Gram-negative systems can deliver substrates, including proteins (green dots), to phylogenetically diverse target cells (–80).
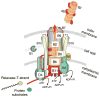
Fig. 2
DNA uptake during transformation in B. subtilis. The uptake machinery is preferentially located at the cell poles. The Ψ-prepilins are processed by the peptidase and translocate to the outer face of the membrane. With the aid of the other ComG proteins, the major Ψ-pilin ComGC assembles into the Ψ-pilus, which attaches exogenous DNA via a hypothetical DNA binding protein. Retraction of the Ψ-pilus, driven by the proton motive force, and DNA binding to the receptor (ComEA) are required to transport one strand of DNA through the membrane channel (ComEC) while the other is degraded by an unidentified nuclease. The helicase/DNA translocase (ComFA) assists the process, along with ssDNA binding proteins that interact with the incoming DNA. RecA forms a filament around the ssDNA, and mediates a search for homology with chromosomal DNA. ADP, adenosine diphosphate; Pi, inorganic phosphate; PMF, proton motive force; ssb, single-stranded DNA binding protein.
Fig. 3
Conjugative DNA transfer through the A. tumefaciens VirB/D4 system. DNA and protein substrates dock initially at the VirD4 receptor, then transfer in succession to the channel components VirB11 ATPase, VirB6, and VirB8, and finally VirB2 and VirB9. Three ATPases (VirD4, VirB4, VirB11) energize DNA substrate transfer through the membrane translocase comprised of either or both VirD4 and VirB6. The DNA substrate translocates to the cell surface via a channel comprised of VirB2 pilin and secretin-like VirB9. ATP energy also induces a structural transition (double-ended arrow) in VirB10 to mediate substrate transfer to the distal portion of the secretion channel.
Similar articles
- Membrane-associated DNA transport machines.
Burton B, Dubnau D. Burton B, et al. Cold Spring Harb Perspect Biol. 2010 Jul;2(7):a000406. doi: 10.1101/cshperspect.a000406. Epub 2010 Jun 23. Cold Spring Harb Perspect Biol. 2010. PMID: 20573715 Free PMC article. Review. - Efficient gene transfer in bacterial cell chains.
Babic A, Berkmen MB, Lee CA, Grossman AD. Babic A, et al. mBio. 2011 Mar 15;2(2):e00027-11. doi: 10.1128/mBio.00027-11. Print 2011. mBio. 2011. PMID: 21406598 Free PMC article. - Horizontal DNA transfer between bacteria in the environment.
Wolska KI. Wolska KI. Acta Microbiol Pol. 2003;52(3):233-43. Acta Microbiol Pol. 2003. PMID: 14743976 Review. - [Horizontal transfer of bacterial genes and its significance for antibiotic resistance and pathogenicity].
Smeets LC, Vandenbroucke-Grauls CM. Smeets LC, et al. Ned Tijdschr Geneeskd. 2007 Dec 8;151(49):2709-14. Ned Tijdschr Geneeskd. 2007. PMID: 18225789 Review. Dutch. - Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells.
Christie PJ, Vogel JP. Christie PJ, et al. Trends Microbiol. 2000 Aug;8(8):354-60. doi: 10.1016/s0966-842x(00)01792-3. Trends Microbiol. 2000. PMID: 10920394 Free PMC article. Review.
Cited by
- Effects of DNA size on transformation and recombination efficiencies in Xylella fastidiosa.
Kung SH, Retchless AC, Kwan JY, Almeida RP. Kung SH, et al. Appl Environ Microbiol. 2013 Mar;79(5):1712-7. doi: 10.1128/AEM.03525-12. Epub 2013 Jan 11. Appl Environ Microbiol. 2013. PMID: 23315739 Free PMC article. - Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins.
Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC. Yadav T, et al. Nucleic Acids Res. 2012 Jul;40(12):5546-59. doi: 10.1093/nar/gks173. Epub 2012 Feb 28. Nucleic Acids Res. 2012. PMID: 22373918 Free PMC article. - Abiotic Gene Transfer: Rare or Rampant?
Kotnik T, Weaver JC. Kotnik T, et al. J Membr Biol. 2016 Oct;249(5):623-631. doi: 10.1007/s00232-016-9897-y. Epub 2016 Apr 11. J Membr Biol. 2016. PMID: 27067073 Free PMC article. Review. - Antimicrobial resistance in the food chain: a review.
Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen MA, Van Huffel X, Imberechts H, Dierick K, Daube G, Saegerman C, De Block J, Dewulf J, Herman L. Verraes C, et al. Int J Environ Res Public Health. 2013 Jun 28;10(7):2643-69. doi: 10.3390/ijerph10072643. Int J Environ Res Public Health. 2013. PMID: 23812024 Free PMC article. Review. - Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient.
Palchevskiy V, Finkel SE. Palchevskiy V, et al. J Bacteriol. 2006 Jun;188(11):3902-10. doi: 10.1128/JB.01974-05. J Bacteriol. 2006. PMID: 16707682 Free PMC article.
References
- Errington J, Bath J, Wu LJ. Nat Rev Mol Cell Biol. 2001;2:538. - PubMed
- Redfield RJ. J Hered. 1993;84:400. - PubMed
- Burrus V, Waldor MK. Res Microbiol. 2004;155:376. - PubMed
- Hofreuter D, Odenbreit S, Haas R. Mol Microbiol. 2001;41:379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM057720/GM/NIGMS NIH HHS/United States
- GM48746/GM/NIGMS NIH HHS/United States
- R01 GM043756/GM/NIGMS NIH HHS/United States
- R01 GM048746-14/GM/NIGMS NIH HHS/United States
- GM43756/GM/NIGMS NIH HHS/United States
- R01 GM048746/GM/NIGMS NIH HHS/United States
- R01 GM057720/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials