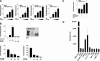NIR is a novel INHAT repressor that modulates the transcriptional activity of p53 - PubMed (original) (raw)
. 2005 Dec 1;19(23):2912-24.
doi: 10.1101/gad.351205.
Natalia Kunowska, Ulrich P Mayer, Judith M Müller, Kristina Heyne, Na Yin, Claudia Fritzsche, Cecilia Poli, Laurent Miguet, Ingo W Schupp, Leo A van Grunsven, Noëlle Potiers, Alain van Dorsselaer, Eric Metzger, Klaus Roemer, Roland Schüle
Affiliations
- PMID: 16322561
- PMCID: PMC1315397
- DOI: 10.1101/gad.351205
NIR is a novel INHAT repressor that modulates the transcriptional activity of p53
Philip Hublitz et al. Genes Dev. 2005.
Abstract
Most transcriptional repression pathways depend on the targeted deacetylation of histone tails. In this report, we characterize NIR, a novel transcriptional corepressor with inhibitor of histone acetyltransferase (INHAT) activity. NIR (Novel INHAT Repressor) is ubiquitously expressed throughout embryonic development and adulthood. NIR is a potent transcriptional corepressor that is not blocked by histone deacetylase inhibitors and is capable of silencing both basal and activator-driven transcription. NIR directly binds to nucleosomes and core histones and prevents acetylation by histone acetyltransferases, thus acting as a bona fide INHAT. Using a tandem affinity purification approach, we identified the tumor suppressor p53 as a NIR-interacting partner. Association of p53 and NIR was verified in vitro and in vivo. Upon recruitment by p53, NIR represses transcription of both p53-dependent reporters and endogenous target genes. Knock-down of NIR by RNA interference significantly enhances histone acetylation at p53-regulated promoters. Moreover, p53-dependent apoptosis is robustly increased upon depletion of NIR. In summary, our findings describe NIR as a novel INHAT that plays an important role in the control of p53 function.
Figures
Figure 1.
Schematic representation and sequence of human NIR. (Top) Schematic representation of the human NIR protein. (Bottom) Amino acid sequence and functional motifs in NIR. INHAT regions are shaded in black. Set/TAF1β homology regions are underlined white. The putative bipartite nuclear localization signal (NLS) is in gray.
Figure 2.
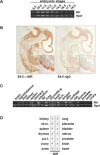
NIR expression analyses. (A) Expression of Nir during mouse embryogenesis. RT-PCR analyses were performed at the indicated stages from E8.5 to E18.5. (B, left) Expression of Nir in sagittal sections of E9.5 mice. Nir is expressed throughout the embryo. Higher magnification exemplifies Nir expression in the somites. The right panel displays staining with a control antibody. (C) Nir expression in tissues of adult mice as determined by RT-PCR. (D) Expression of NIR in human adult tissues. mRNA dot-blot hybridization shows that NIR is ubiquitously expressed in human adult tissues. (p.b.l.) Peripheral blood lymphocytes, (sk.m.) skeletal muscle, (thyroid/p.g.) thyroid and parathyroid gland.
Figure 3.
NIR is an inhibitor of histone acetylation. (A) NIR interacts with core histones and nucleosomes in vitro. Bacterially expressed GST-NIR INHAT domains interact with core histones (left panel) and nucleosomes (right panel). The unrelated protein Nix1 serves as a control. Proteins are visualized by Coomassie staining. (B) [35S]methionine-labeled in vitro translated NIR interacts with nonacetylated Sepharose-coupled peptides corresponding to the N-terminal tail of histone H3 (amino acids 1-30). Acetylation at K9, K14, K18, or K23 no longer permits binding of NIR. Interaction of radioactively labeled NIR with histone tails is visualized by autoradiography. (C) NIR is a functional INHAT. (Left panel) Bacterially expressed NIR INHAT regions prevent acetylation of all core histones by the p300 histone acetyltransferase in vitro. To demonstrate specificity, the unrelated protein Nix1 is used as a control. INHAT activity is visualized by autoradiography. (Right panel) The Coomassie stain verifies the use of equal amounts of His-tagged proteins and absence of protease contaminations that could potentially degrade histones. (D) Full-length NIR-TAP and the indicated deletion mutants were tested for association with nucleosomes and INHAT activity. (Upper panel) Equal amounts of TAP fusion proteins are visualized by Western blotting. (Middle panel) Pulldown of NIR-TAP proteins with nucleosomes. The lower panel displays the INHAT assays using either core histones or nucleosomes as substrates and full-length NIR-TAP or the indicated mutants. Full-length NIR and both INHAT-domains block acetylation by the p300. INHAT activity is visualized by autoradiography. Ponceau S staining verifies the presence of equal amounts of nucleosomes and excludes protease contamination of affinity-purified NIR proteins.
Figure 4.
NIR is a potent HDAC-independent transcriptional repressor. (A) Gal-NIR represses transcription of synthetic minimal or complex Gal-dependent reporters in a dose-dependent manner in various cell lines. In transient transfection assays, 50, 100, or 500 ng of Gal-NIR expression plasmid were used. As a control, the corresponding amounts of Gal4-DBD (Gal) were transfected. (B) NIR represses activated transcription. BHK cells were cotransfected with 500 ng of L8G5-LUC reporter and 100 ng of LexA-VP16, Gal-CREB, or Gal-TIF2 expression plasmids. Fold activation is calculated in relation to the corresponding amounts of Gal4-DBD. (C) NIR repression is not influenced by HDAC inhibitors. BHK cells were cotransfected with 500 ng of Gal-TK-LUC reporter and 50 ng of Gal-NIR expression plasmid. Na-butyrate (5 mM), nicotinamide (10 mM), and TSA (330 nM) were applied for 24 h post-transfection. Fold repression is calculated in relation to the corresponding amounts of Gal4-DBD. (D) NIR is not associated with HDAC activity. NIR and associated complexes were immunopurified from 293 cells with an α-NIR (2719) antibody. Release of radiolabeled acetyl groups from H3 peptides is observed only when using whole-cell extract (w.c.e.) or purified N-CoR-associated complexes. The amount of released radiolabeled acetyl groups (measured by β-counting) in NIR immunoprecipitates corresponds to the mock-treated reactions or to the release control. Specificity of the reactions is demonstrated by using the HDAC-inhibitor TSA. Bars represent mean ± SD (n ≥ 6).
Figure 5.
NIR interacts with p53 in vitro and in vivo. (A) NIR is a nuclear protein and colocalizes with p53 in vivo in BHK cells. NIR immunoreactivity is displayed in green, p53 in red. Nuclei were stained with DAPI (blue). For clarity, the merge image displays only NIR and p53 immunoreactivity. (B) p53 and NIR interact in vitro. Interaction domains were mapped in GST-pulldown assays using GST-p53, GST-p53 deletion mutants, and [35S]methionine-labeled in vitro translated NIR or mutants thereof. The use of equal amounts of GST fusion proteins is demonstrated by Coomassie staining. (C) Coimmunoprecipitation of TAP-tagged NIR and myc-tagged p53 in BHK cells. Membranes were decorated with the α-NIR 2910 or the α-p53 DO-1 antibody. Five percent of whole-cell extract was used as input. (D) Endogenous p53 coimmunoprecipitates with NIR. Whole-cell extracts from 293 cells and nuclear extracts from HCT116/p53+/+ cells mock-treated or treated with doxorubicin (DOX) were immunoprecipitated with the α-NIR 2910 antibody. Membranes were decorated with α-NIR (2910) or α-p53 (DO-1) antibody. We used 2.5% and 25% of whole-cell extract as inputs for p53 and NIR, respectively.
Figure 6.
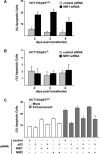
NIR regulates p53 function. (A) NIR represses p53 transcriptional activity. Transient transfections were carried out in the p53-negative cell lines Saos-2 and HCT116/p53-/-. NIR blocks p53-induced expression of the synthetic p53-responsive reporter PG13, whereas the control reporter with mutated p53-binding sites (MG15) is not influenced by either p53 or NIR. NIR represses activation of the known p53 target genes p21, PIG3, and Noxa. (RLU) Relative light units. Bars represent mean ± SD (n ≥ 6). (B) NIR mediates repression of endogenous p53 target genes. Mock-treated or doxorubicin (DOX)-stressed HCT116/p53+/+ and HCT116/p53-/- cells were transfected with either a Flag-control plasmid or with a plasmid coding for Flag-NIR. RT-PCR analyses were performed with primers amplifying the p53 target genes PIG3, NOXA, and p21. Amplification of the housekeeping gene GAPDH serves as a control. (C) NIR and p53 are present at endogenous p53-regulated promoters. ChIP assays show that NIR is localized at p53 target promoters in HCT116/p53-/- cells only in the presence of transfected p53. The p53 target genes p21 and PIG3 are precipitated using α-NIR (2910) or α-p53 (DO-7) antibody. Unrelated promoters (U6 and GAPDH) are not enriched. (D) Endogenously expressed NIR and p53 are present at p53-regulated promoters in HCT116p53+/+ cells. In ChIP assays the p21 promoter is precipitated using antibodies directed against either NIR (2910) or p53 (DO-7). The unrelated U6 and GAPDH promoters are not enriched. (E, upper panel) Re-ChIP confirms that NIR and p53 form a complex on the endogenous p21 promoter. First ChIP was performed using the α-NIR 2910 antibody. Re-ChIP was performed using either mIgG or the α-p53 DO-1 antibody. The unrelated U6 promoter is not precipitated by the antibodies. (Lower panel) NIR and p53 are present at the endogenous p21 promoter in HCT116p53+/+ cells in the absence (-) or presence (+) of doxorubicin-induced cellular stress. (F) siRNA-mediated NIR knock-down correlates with increased p21 protein and RNA levels in unstressed HCT116/p53+/+ cells. Western blotting and RT-PCR analyses were performed at day 3 post-transfection with siRNA. ACTIN serves as a control for equal protein loading. (G) NIR acts as a p53-recruited INHAT in vivo. Knock-down of NIR by siRNA (NIR1) in HCT116/p53+/+ cells leads to histone hyperacetylation at p53 target promoters as shown by ChIP assays using a combination of α-acetylated H3 and H4 antibodies. Unrelated promoters (U6 and GAPDH) are not enriched. The control ChIP from HCT116/p53-/- cells did not reveal changes in the acetylation status of the p21 promoter, thus demonstrating specificity.
Figure 7.
Knock-down of NIR increases p53-dependent apoptosis. NIR knock-down in HCT116/p53+/+ cells results in a strong apoptotic response. p53-containing (A) and p53-deficient (B) HCT116 cells were transfected with either a control siRNA or a siRNA directed against NIR (NIR1). The percentage of apoptotic cells was determined at the indicated time points after transfection. (C) Apoptosis induced by NIR knock-down is p53-dependent. HCT116/p53+/+ cells were transfected with either an unrelated siRNA (control) or the indicated combinations of siRNAs directed against p53 or NIR (NIR1 and NIR2). Apoptosis was assessed under unstressed or stress-induced (5-fluorouracil) conditions. Bars represent mean ± SD (n ≥ 6).
Similar articles
- Deacetylation of p53 modulates its effect on cell growth and apoptosis.
Luo J, Su F, Chen D, Shiloh A, Gu W. Luo J, et al. Nature. 2000 Nov 16;408(6810):377-81. doi: 10.1038/35042612. Nature. 2000. PMID: 11099047 - Tumor-specific adenoviral gene therapy: transcriptional repression of gene expression by utilizing p53-signal transduction pathways.
Kühnel F, Zender L, Wirth T, Schulte B, Trautwein C, Manns M, Kubicka S. Kühnel F, et al. Cancer Gene Ther. 2004 Jan;11(1):28-40. doi: 10.1038/sj.cgt.7700632. Cancer Gene Ther. 2004. PMID: 14681724 - Regulation of the p53 transcriptional activity.
Liu G, Chen X. Liu G, et al. J Cell Biochem. 2006 Feb 15;97(3):448-58. doi: 10.1002/jcb.20700. J Cell Biochem. 2006. PMID: 16288459 Review. - Methods to study p53-repressed promoters.
Dumont P, Della Pietra A, Murphy ME. Dumont P, et al. Methods Mol Biol. 2003;234:111-20. doi: 10.1385/1-59259-408-5:111. Methods Mol Biol. 2003. PMID: 12824528 - Acetylation of non-histone proteins modulates cellular signalling at multiple levels.
Spange S, Wagner T, Heinzel T, Krämer OH. Spange S, et al. Int J Biochem Cell Biol. 2009 Jan;41(1):185-98. doi: 10.1016/j.biocel.2008.08.027. Epub 2008 Sep 2. Int J Biochem Cell Biol. 2009. PMID: 18804549 Review.
Cited by
- Aurora B interacts with NIR-p53, leading to p53 phosphorylation in its DNA-binding domain and subsequent functional suppression.
Wu L, Ma CA, Zhao Y, Jain A. Wu L, et al. J Biol Chem. 2011 Jan 21;286(3):2236-44. doi: 10.1074/jbc.M110.174755. Epub 2010 Oct 19. J Biol Chem. 2011. PMID: 20959462 Free PMC article. - REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana.
Prunet N, Morel P, Thierry AM, Eshed Y, Bowman JL, Negrutiu I, Trehin C. Prunet N, et al. Plant Cell. 2008 Apr;20(4):901-19. doi: 10.1105/tpc.107.053306. Epub 2008 Apr 25. Plant Cell. 2008. PMID: 18441215 Free PMC article. - Susceptibility loci for pancreatic cancer in the Brazilian population.
Aoki MN, Stein A, de Oliveira JC, Chammas R, Uno M, Munhoz FBA, Marin AM, Canzian F. Aoki MN, et al. BMC Med Genomics. 2021 Apr 20;14(1):111. doi: 10.1186/s12920-021-00956-5. BMC Med Genomics. 2021. PMID: 33879152 Free PMC article. - The role of LANP and ataxin 1 in E4F-mediated transcriptional repression.
Cvetanovic M, Rooney RJ, Garcia JJ, Toporovskaya N, Zoghbi HY, Opal P. Cvetanovic M, et al. EMBO Rep. 2007 Jul;8(7):671-7. doi: 10.1038/sj.embor.7400983. Epub 2007 Jun 8. EMBO Rep. 2007. PMID: 17557114 Free PMC article. - LncRNA RUS shapes the gene expression program towards neurogenesis.
Schneider MF, Müller V, Müller SA, Lichtenthaler SF, Becker PB, Scheuermann JC. Schneider MF, et al. Life Sci Alliance. 2022 Jun 10;5(10):e202201504. doi: 10.26508/lsa.202201504. Print 2022 Oct. Life Sci Alliance. 2022. PMID: 35688487 Free PMC article.
References
- Allison, S.J. and Milner, J. 2004. Remodeling chromatin on a global scale: A novel protective function of p53. Carcinogenesis 25: 1551-1557. - PubMed
- Choi, Y.B., Ko, J.K., and Shin, J. 2004. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J. Biol. Chem. 279: 50930-50941. - PubMed
- Christian, M., Tullet, J.M., and Parker, M.G. 2004. Characterization of four autonomous repression domains in the corepressor receptor interacting protein 140. J. Biol. Chem. 279: 15645-15651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous