A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia - PubMed (original) (raw)
Case Reports
. 2005 Dec;115(12):3506-16.
doi: 10.1172/JCI24832.
Agnes van Horssen-Zoetbrood, Jeffrey M Beekman, Britt Otterud, Frans Maas, Rob Woestenenk, Michel Kester, Mark Leppert, Anton V Schattenberg, Theo de Witte, Elly van de Wiel-van Kemenade, Harry Dolstra
Affiliations
- PMID: 16322791
- PMCID: PMC1297240
- DOI: 10.1172/JCI24832
Case Reports
A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia
Björn de Rijke et al. J Clin Invest. 2005 Dec.
Abstract
Minor histocompatibility antigens (mHAgs) constitute the targets of the graft-versus-leukemia response after HLA-identical allogeneic stem cell transplantation. Here, we have used genetic linkage analysis to identify a novel mHAg, designated lymphoid-restricted histocompatibility antigen-1 (LRH-1), which is encoded by the P2X5 gene and elicited an allogeneic CTL response in a patient with chronic myeloid leukemia after donor lymphocyte infusion. We demonstrate that immunogenicity for LRH-1 is due to differential protein expression in recipient and donor cells as a consequence of a homozygous frameshift polymorphism in the donor. Tetramer analysis showed that emergence of LRH-1-specific CD8+ cytotoxic T cells in peripheral blood and bone marrow correlated with complete remission of chronic myeloid leukemia. Furthermore, the restricted expression of LRH-1 in hematopoietic cells including leukemic CD34+ progenitor cells provides evidence of a role for LRH-1-specific CD8+ cytotoxic T cells in selective graft-versus-leukemia reactivity in the absence of severe graft-versus-host disease. These findings illustrate that the P2X5-encoded mHAg LRH-1 could be an attractive target for specific immunotherapy to treat hematological malignancies recurring after allogeneic stem cell transplantation.
Figures
Figure 1
Specific reactivity of HLA-B*0702–restricted CTL RP1 against hematopoietic cells. (A) Specific cytotoxicity was tested in 51Cr release assays against EBV-LCLs of the recipient (Rt) and donor (Do). NK-sensitive K562 cells were used to determine nonspecific lysis. Effector to target (E/T) ratios are indicated. (B) Production of IFN-γ by CTL RP1 stimulated with recipient EBV-LCLs, EBV-LCLs of 2 unrelated individuals (nos. 1 and 2) sharing HLA-B7 with the recipient, and an EBV-LCL of an HLA class I–mismatched individual (no. 3) that was transduced with HLA-B*0702. Data are displayed as mean IFN-γ release ± SD of triplicate wells. (C) Specific cytotoxicity against hematopoietic cells of lymphoid origin (EBV-LCLs, PHA-stimulated T cells, and CD40L-stimulated B cells) and BM-derived fibroblasts incubated with 10 ng/ml TNF-α and 100 U/ml IFN-γ for 2 days before 51Cr labeling. The E/T ratio was 1:1. (D) IFN-γ production by CTL RP1 upon stimulation with hematopoietic cells of myeloid origin (monocytes, immature DCs [iDC] and mature DCs [mDC]).
Figure 2
The LRH-1 locus is closely linked to a cluster of markers on chromosome 17p13.2. (A) LRH-1 segregation pattern for individuals of the CEPH pedigrees 1331, 1332, 1347, 1413, 1362, and 102. EBV-LCLs of all available family members were transduced with HLA-B*0702 and tested for recognition by CTL RP1. Filled circles (females) or squares (males) represent individuals scored as positive for the LRH-1 phenotype, and open circles (females) or squares (males) represent individuals scored as negative. Shaded symbols represent individuals from whom no EBV-LCLs was available. (B) Genetic map of chromosome 17p13.2 showing relative marker loci oriented with the centromere at the bottom of the figure. lod scores summed for all available families used in the linkage analysis are shown to the right of each genomic marker. CEPH families that are not genotyped for a particular marker and therefore not included in the linkage analysis are indicated in parentheses.
Figure 3
Identification of P2X5 as the gene encoding the HLA-B*0702–restricted epitope LRH-1. (A) IFN-γ production by CTL RP1 upon stimulation with EBV-LCL from the homozygous LRH-1–positive individual 1331-8234 and 293T-HLA-B*0702 cells transfected with full-length constructs of P2X5 transcript variants 1 and 2, and deletion constructs of P2X5 transcript variant 1 encoding the first 5 exons (Var 1 Δ5) either generated from EBV-LCL of the homozygous LRH-1–positive individual 1331–8234 or the homozygous LRH-1–negative stem cell donor. (B) Schematic representation of the P2X5 gene on chromosome 17p13.2. The nucleotide and deduced aa sequences of exons 3 and 4 of both the LRH-1–positive and –negative allele present in recipient and donor, respectively, are shown. Disparity between the recipient and donor P2X5 protein sequence is due to a frameshift induced by a cytosine deletion polymorphism in the nucleotide sequence of exon 3 (5C versus 4C). This results in a disparate aa sequence of 9 residues in the recipient direction (shaded box) and 35 residues in the donor direction (open box). The aa sequence of the antigenic epitope that is recognized by the HLA-B*0702–restricted CTL RP1 is underlined. (C) LRH-1 epitope reconstitution with synthetic peptides corresponding to the P2X5 exon 3 sequence of the recipient. Donor EBV-LCL was pulsed with various peptide concentrations and tested for recognition by CTL RP1. IFN-γ production was determined by ELISA.
Figure 4
Detection of LRH-1–specific CD8+ T cells in peripheral blood of CML patient UPN389 after DLI. PBMCs collected 3, 6, 12, and 38 months after DLI-2 were stained with LRH-1/HLA-B7 tetramer, anti-CD8, anti-CD45, anti-CD3, and 7-amino-actinomycin D (7AAD). Subsequently, cell populations were analyzed by flow cytometry. Cells were gated on CD45+CD3+7AAD– lymphocytes, and the percentages of tetramer-binding cells among CD8+ T cells are given. The remaining PBMCs were stimulated twice with LRH-1 peptide–pulsed (10 μM) EBV-LCL of the donor and assayed on day 14 for LRH-1 tetramer-binding CD8+ T cells.
Figure 5
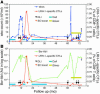
Longitudinal follow-up of LRH-1–specific CD8+ T cells in peripheral blood from CML patient UPN389 in relation to clinical outcome. (A) The percentage of LRH-1 tetramer-positive cells among CD8+ T cells (right y axis) are compared with the wbc count × 106 per ml peripheral blood (left y axis). (B) The percentages of LRH-1 tetramer-positive cells among CD8+ T cells (right y axis) are compared with the relative units of Bcr-Abl in peripheral blood (left y axis). The lower level of detection of Bcr-Abl PCR is 4 × 10–6, depicted by the dotted line. Administration of DLIs 1–5 and stem cell (SC) boost are indicated by black and orange arrows, respectively. Treatment intervals with CsA and imatinib mesylate (Glivec) are shown by blue and yellow bars, respectively.
Figure 6
P2X5 gene expression is restricted to leukemic and normal CD34+ progenitor cells as well as lymphoid cells. (A) P2X5 expression determined by real-time quantitative PCR in CD34+ subpopulations isolated from leukemia patients (CML blast crises, n = 4 and acute myeloid leukemia, n = 10) and healthy stem cell donors (normal BM, n = 4 and G-CSF–mobilized peripheral blood, n = 5). Leukemic CD34+ subsets isolated from CML patient UPN389 at first relapse are indicated by the arrows. (B) P2X5 expression determined by real-time quantitative RT-PCR in freshly isolated cells or primary cell cultures of hematopoietic and nonhematopoietic origin. Expression is shown relative to the P2X5 expression measured in the reference cell line JVM-2, which is susceptible to lysis by the LRH-1–specific CTL RP1. The housekeeping Pbgd gene was used for normalization. Cell types with P2X5 expression less than 0.4 were not recognized by CTL RP1, indicating that these cell types can be considered as LRH-1 negative. This arbitrary threshold is indicated with a dashed line. The mean expression level for each cell population is shown by the thick line.
Comment in
- Expanding the immunotherapeutic potential of minor histocompatibility antigens.
Spierings E, Goulmy E. Spierings E, et al. J Clin Invest. 2005 Dec;115(12):3397-400. doi: 10.1172/JCI27094. J Clin Invest. 2005. PMID: 16322786 Free PMC article.
Similar articles
- Efficient activation of LRH-1-specific CD8+ T-cell responses from transplanted leukemia patients by stimulation with P2X5 mRNA-electroporated dendritic cells.
Overes IM, Fredrix H, Kester MG, Falkenburg JH, van der Voort R, de Witte TM, Dolstra H. Overes IM, et al. J Immunother. 2009 Jul-Aug;32(6):539-51. doi: 10.1097/CJI.0b013e3181987c22. J Immunother. 2009. PMID: 19483655 - Expression of P2X5 in lymphoid malignancies results in LRH-1-specific cytotoxic T-cell-mediated lysis.
Overes IM, de Rijke B, van Horssen-Zoetbrood A, Fredrix H, de Graaf AO, Jansen JH, van Krieken JH, Raymakers RA, van der Voort R, de Witte TM, Dolstra H. Overes IM, et al. Br J Haematol. 2008 Jun;141(6):799-807. doi: 10.1111/j.1365-2141.2008.07125.x. Epub 2008 Apr 10. Br J Haematol. 2008. PMID: 18410452 - Myeloid leukemic progenitor cells can be specifically targeted by minor histocompatibility antigen LRH-1-reactive cytotoxic T cells.
Norde WJ, Overes IM, Maas F, Fredrix H, Vos JC, Kester MG, van der Voort R, Jedema I, Falkenburg JH, Schattenberg AV, de Witte TM, Dolstra H. Norde WJ, et al. Blood. 2009 Mar 5;113(10):2312-23. doi: 10.1182/blood-2008-04-153825. Epub 2008 Dec 12. Blood. 2009. PMID: 19074734 - Molecular basis for therapeutic decisions in chronic myeloid leukemia patients after allogeneic bone marrow transplantation.
Román J, Alvarez MA, Torres A. Román J, et al. Haematologica. 2000 Oct;85(10):1072-82. Haematologica. 2000. PMID: 11025600 Review. - Cytotoxic T-lymphocyte (CTL) responses against acute or chronic myeloid leukemia.
Falkenburg JH, Smit WM, Willemze R. Falkenburg JH, et al. Immunol Rev. 1997 Jun;157:223-30. doi: 10.1111/j.1600-065x.1997.tb00985.x. Immunol Rev. 1997. PMID: 9255633 Review.
Cited by
- Leukemia-associated minor histocompatibility antigen discovery using T-cell clones isolated by in vitro stimulation of naive CD8+ T cells.
Bleakley M, Otterud BE, Richardt JL, Mollerup AD, Hudecek M, Nishida T, Chaney CN, Warren EH, Leppert MF, Riddell SR. Bleakley M, et al. Blood. 2010 Jun 10;115(23):4923-33. doi: 10.1182/blood-2009-12-260539. Epub 2010 Mar 4. Blood. 2010. PMID: 20203263 Free PMC article. - Exploiting T cells specific for human minor histocompatibility antigens for therapy of leukemia.
Bleakley M, Riddell SR. Bleakley M, et al. Immunol Cell Biol. 2011 Mar;89(3):396-407. doi: 10.1038/icb.2010.124. Epub 2011 Feb 8. Immunol Cell Biol. 2011. PMID: 21301477 Free PMC article. Review. - Rehabilitation of the P2X5 receptor: a re-evaluation of structure and function.
King BF. King BF. Purinergic Signal. 2023 Jun;19(2):421-439. doi: 10.1007/s11302-022-09903-0. Epub 2022 Oct 24. Purinergic Signal. 2023. PMID: 36279087 Free PMC article. Review. - Molecular properties of P2X receptors.
Roberts JA, Vial C, Digby HR, Agboh KC, Wen H, Atterbury-Thomas A, Evans RJ. Roberts JA, et al. Pflugers Arch. 2006 Aug;452(5):486-500. doi: 10.1007/s00424-006-0073-6. Epub 2006 Apr 11. Pflugers Arch. 2006. PMID: 16607539 Review. - Moderate exercise increases expression for sensory, adrenergic, and immune genes in chronic fatigue syndrome patients but not in normal subjects.
Light AR, White AT, Hughen RW, Light KC. Light AR, et al. J Pain. 2009 Oct;10(10):1099-112. doi: 10.1016/j.jpain.2009.06.003. Epub 2009 Jul 31. J Pain. 2009. PMID: 19647494 Free PMC article.
References
- Horowitz MM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. - PubMed
- Radich JP, Olavarria E, Apperley JF. Allogeneic hematopoietic stem cell transplantation for chronic myeloid leukemia. Hematol. Oncol. Clin. North Am. 2004;18:685–702. - PubMed
- Sucia S, et al. Allogeneic compared with autologous stem cell transplantation in the treatment of patients younger than 46 years with acute myeloid leukemia (AML) in first complete remission (CR1): an intention-to-treat analysis of the EORTC/GIMEMAAML-10 trial. Blood. 2003;102:1232–1240. - PubMed
- Goulmy E. Human minor histocompatibility antigens: new concepts for marrow transplantation and adoptive immunotherapy. Immunol. Rev. 1997;157:125–140. - PubMed
- Bleakley M, Riddell SR. Molecules and mechanisms of the graft-versus-leukaemia effect. Nat. Rev. Cancer. 2004;4:371–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials