Medical sequencing of candidate genes for nonsyndromic cleft lip and palate - PubMed (original) (raw)
doi: 10.1371/journal.pgen.0010064. Epub 2005 Dec 2.
Joseph R Avila, Sandra Daack-Hirsch, Ecaterina Dragan, Têmis M Félix, Fedik Rahimov, Jill Harrington, Rebecca R Schultz, Yoriko Watanabe, Marla Johnson, Jennifer Fang, Sarah E O'Brien, Iêda M Orioli, Eduardo E Castilla, David R Fitzpatrick, Rulang Jiang, Mary L Marazita, Jeffrey C Murray
Affiliations
- PMID: 16327884
- PMCID: PMC1298935
- DOI: 10.1371/journal.pgen.0010064
Medical sequencing of candidate genes for nonsyndromic cleft lip and palate
Alexandre R Vieira et al. PLoS Genet. 2005 Dec.
Abstract
Nonsyndromic or isolated cleft lip with or without cleft palate (CL/P) occurs in wide geographic distribution with an average birth prevalence of 1/700. We used direct sequencing as an approach to study candidate genes for CL/P. We report here the results of sequencing on 20 candidate genes for clefts in 184 cases with CL/P selected with an emphasis on severity and positive family history. Genes were selected based on expression patterns, animal models, and/or role in known human clefting syndromes. For seven genes with identified coding mutations that are potentially etiologic, we performed linkage disequilibrium studies as well in 501 family triads (affected child/mother/father). The recently reported MSX1 P147Q mutation was also studied in an additional 1,098 cleft cases. Selected missense mutations were screened in 1,064 controls from unrelated individuals on the Centre d'Etude du Polymorphisme Humain (CEPH) diversity cell line panel. Our aggregate data suggest that point mutations in these candidate genes are likely to contribute to 6% of isolated clefts, particularly those with more severe phenotypes (bilateral cleft of the lip with cleft palate). Additional cases, possibly due to microdeletions or isodisomy, were also detected and may contribute to clefts as well. Sequence analysis alone suggests that point mutations in FOXE1, GLI2, JAG2, LHX8, MSX1, MSX2, SATB2, SKI, SPRY2, and TBX10 may be rare causes of isolated cleft lip with or without cleft palate, and the linkage disequilibrium data support a larger, as yet unspecified, role for variants in or near MSX2, JAG2, and SKI. This study also illustrates the need to test large numbers of controls to distinguish rare polymorphic variants and prioritize functional studies for rare point mutations.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Protein Comparisons of the Available Gene Orthologs for GLI2 S1213Y and SPRY2 D20A
GLI2 S1213Y (A) and SPRY2 D20A (B): Green bars indicate degree of conservation in each site. Amino acids in red indicate the mutation sites. All mutation comparisons are available as supplemental material at
http://genetics.uiowa.edu/publications.html
.
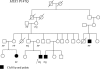
Figure 2. Family Pedigree of the Filipino Family Segregating the MSX1 P147Q Mutation
PP (proline–proline) indicates the wild-type genotype. PQ (proline–glutamine) indicates the genotype of the individuals carrying the mutation.
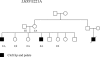
Figure 3. Family Pedigree of the Filipino Proband with the LHX8 E221A Mutation
The segregation pattern of the mutation is not consistent with a simple Mendelian model for the disease. EE (glutamic acid–glutamic acid) indicates the wild-type genotype. EA (glutamic acid–alanine) indicates the genotype of the individuals carrying the mutation.
Similar articles
- Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate.
Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, Romitti PA, Christensen K, Orioli IM, Castilla EE, Machida J, Natsume N, Murray JC. Jezewski PA, et al. J Med Genet. 2003 Jun;40(6):399-407. doi: 10.1136/jmg.40.6.399. J Med Genet. 2003. PMID: 12807959 Free PMC article. - Targeted scan of fifteen regions for nonsyndromic cleft lip and palate in Filipino families.
Schultz RE, Cooper ME, Daack-Hirsch S, Shi M, Nepomucena B, Graf KA, O'Brien EK, O'Brien SE, Marazita ML, Murray JC. Schultz RE, et al. Am J Med Genet A. 2004 Feb 15;125A(1):17-22. doi: 10.1002/ajmg.a.20424. Am J Med Genet A. 2004. PMID: 14755461 - Mutation analysis of the MSX1 gene exons and intron in patients with nonsyndromic cleft lip and palate.
Lace B, Vasiljeva I, Dundure I, Barkane B, Akota I, Krumina A. Lace B, et al. Stomatologija. 2006;8(1):21-4. Stomatologija. 2006. PMID: 16687911 - Progress toward discerning the genetics of cleft lip.
Lidral AC, Moreno LM. Lidral AC, et al. Curr Opin Pediatr. 2005 Dec;17(6):731-9. doi: 10.1097/01.mop.0000185138.65820.7f. Curr Opin Pediatr. 2005. PMID: 16282779 Free PMC article. Review. - Candidate genes for nonsyndromic cleft lip and palate.
Vieira AR, Orioli IM. Vieira AR, et al. ASDC J Dent Child. 2001 Jul-Aug;68(4):272-9, 229. ASDC J Dent Child. 2001. PMID: 11862881 Review.
Cited by
- Evaluating rare coding variants as contributing causes to non-syndromic cleft lip and palate.
Leslie EJ, Murray JC. Leslie EJ, et al. Clin Genet. 2013 Nov;84(5):496-500. doi: 10.1111/cge.12018. Epub 2012 Oct 10. Clin Genet. 2013. PMID: 22978696 Free PMC article. - Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2.
Cesario JM, Landin Malt A, Deacon LJ, Sandberg M, Vogt D, Tang Z, Zhao Y, Brown S, Rubenstein JL, Jeong J. Cesario JM, et al. Hum Mol Genet. 2015 Sep 1;24(17):5024-39. doi: 10.1093/hmg/ddv223. Epub 2015 Jun 12. Hum Mol Genet. 2015. PMID: 26071365 Free PMC article. - Further delineation of the SATB2 phenotype.
Döcker D, Schubach M, Menzel M, Munz M, Spaich C, Biskup S, Bartholdi D. Döcker D, et al. Eur J Hum Genet. 2014 Aug;22(8):1034-9. doi: 10.1038/ejhg.2013.280. Epub 2013 Dec 4. Eur J Hum Genet. 2014. PMID: 24301056 Free PMC article. - Systematic analysis of copy number variants of a large cohort of orofacial cleft patients identifies candidate genes for orofacial clefts.
Conte F, Oti M, Dixon J, Carels CE, Rubini M, Zhou H. Conte F, et al. Hum Genet. 2016 Jan;135(1):41-59. doi: 10.1007/s00439-015-1606-x. Epub 2015 Nov 11. Hum Genet. 2016. PMID: 26561393 Free PMC article. - Investigation of genetic factors underlying typical orofacial clefts: mutational screening and copy number variation.
Simioni M, Araujo TK, Monlleo IL, Maurer-Morelli CV, Gil-da-Silva-Lopes VL. Simioni M, et al. J Hum Genet. 2015 Jan;60(1):17-25. doi: 10.1038/jhg.2014.96. Epub 2014 Nov 13. J Hum Genet. 2015. PMID: 25391604
References
- Mossey PA, Little J. Epidemiology of oral clefts: An international perspective. In: Wyszynski DF, editor. Cleft lip and palate. From origin to treatment. New York: Oxford Press; 2002. pp. 127–158.
- Suzuki Y, Jezewski PA, Machida J, Watanabe Y, Shi M, et al. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet Med. 2004;6:117–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- D43 TW005503/TW/FIC NIH HHS/United States
- R01 ES010876/ES/NIEHS NIH HHS/United States
- R37 DE008559/DE/NIDCR NIH HHS/United States
- 5 D43 TW05503/TW/FIC NIH HHS/United States
- DE16215/DE/NIDCR NIH HHS/United States
- DE08559/DE/NIDCR NIH HHS/United States
- R01 DE016148/DE/NIDCR NIH HHS/United States
- ES10876/ES/NIEHS NIH HHS/United States
- R01 DE008559/DE/NIDCR NIH HHS/United States
- P50 DE016215/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous