SNEV is an evolutionarily conserved splicing factor whose oligomerization is necessary for spliceosome assembly - PubMed (original) (raw)
. 2005 Dec 6;33(21):6868-83.
doi: 10.1093/nar/gki986. Print 2005.
Paul Ajuh, Guido Stadler, Marlies Löscher, Regina Voglauer, Wolfgang Ernst, Janet Chusainow, Frank Eisenhaber, Marion Pokar, Klaus Fortschegger, Martin Grey, Angus I Lamond, Hermann Katinger
Affiliations
- PMID: 16332694
- PMCID: PMC1310963
- DOI: 10.1093/nar/gki986
SNEV is an evolutionarily conserved splicing factor whose oligomerization is necessary for spliceosome assembly
Johannes Grillari et al. Nucleic Acids Res. 2005.
Abstract
We have isolated the human protein SNEV as downregulated in replicatively senescent cells. Sequence homology to the yeast splicing factor Prp19 suggested that SNEV might be the orthologue of Prp19 and therefore might also be involved in pre-mRNA splicing. We have used various approaches including gene complementation studies in yeast using a temperature sensitive mutant with a pleiotropic phenotype and SNEV immunodepletion from human HeLa nuclear extracts to determine its function. A human-yeast chimera was indeed capable of restoring the wild-type phenotype of the yeast mutant strain. In addition, immunodepletion of SNEV from human nuclear extracts resulted in a decrease of in vitro pre-mRNA splicing efficiency. Furthermore, as part of our analysis of protein-protein interactions within the CDC5L complex, we found that SNEV interacts with itself. The self-interaction domain was mapped to amino acids 56-74 in the protein's sequence and synthetic peptides derived from this region inhibit in vitro splicing by surprisingly interfering with spliceosome formation and stability. These results indicate that SNEV is the human orthologue of yeast PRP19, functions in splicing and that homo-oligomerization of SNEV in HeLa nuclear extract is essential for spliceosome assembly and that it might also be important for spliceosome stability.
Figures
Figure 1
Sequence comparison of SNEV and Prp19p. (A) Domain architecture of human SNEV and yeast Prp19p is similar and consists of a UFD2 like box (U-box), a low complexity region (LCR), a coiled coil region (CC), a putative globular domain (?), as well as a charged region (++−+−). (B) Alignment by BLAST shows 23% overall identities (423/504; 23% identities and 42% positives). Highest homology can be seen at the N-terminus, whereas the C-terminus differs.
Figure 2
Human–yeast chimeric SNEV complements the mutant phenotype of MG5128. (A) Overview of SNEV and Prp19 constructs tested in this study. Length indicates the number of amino acids (aa), MW is the molecular weight (kd), (+) indicates ability to grow at the indicated temperature, to sporulate or to perform forward mutability. (−) no growth, (n.d.) not determined. The star in construct d indicates the point mutation. (B) All chimeric constructs introduced into the mutant yeast strain are translated into protein. Western blots after SDS–PAGE of cell-lysates of MG5128 transformed with pYPGE15, containing various chimeric constructs, which were fused to a His6-tag. Detection was performed using anti-TetraHis mouse antibody as primary and peroxidase-conjugated goat-anti-mouse antibody as secondary antibody. Calculated molecular masses [kDa] are indicated. The small bands in lanes 4–6 and 11–14 represent degradation products resulting from the lysis process. (C) Growth at the permissive (30°C) and the non-permissive (37°C) temperature of MG5128, transformed with the various constructs was analysed. All clones showed growth at 30°C, whereas at 37°C only transformants containing wtPrp19p, SNEV[1–66]Prp19p[68–503], Prp19p[68–503] or SNEV[1–91]Prp19p[98–503] were able to form colonies. Sector 1: wt Prp19p (positive control), 2: empty vector control, 3: SNEV, 4: SNEV[1–66]Prp19p[68–503], 5: Prp19p[68–503], 6: SNEV[1–91]Prp19p[98–503], 7: Prp19p[97–503], 8: SNEV[1–205]Prp19p[208–503]. (D) Ability of the various constructs to sporulate on KAC agar plates. MG5128 transformed with the various constructs was incubated at 30°C for one week, vegetative and sporulated cells were counted. Sporulation was calculated as the ratio of sporulated versus vegetative cells. Mean and standard deviation (SD) were calculated from at least three independent experiments. The total number of cells counted is indicated. (E) The human–yeast chimeric protein SNEV[1–66]Prp19[68–503] restores wild-type phenotype regarding UV sensitivity and forward mutability. Survival of the various MG5128 transformants after UV-irradiation (left panel) (266 nm) and ability of the various chimeric constructs to restore wild-type like forward mutability by growth on canavanine containing media after UV-irradiation (right panel). Number of mutants has been calculated as mutants per UV radiation surviving cells.
Figure 2
Human–yeast chimeric SNEV complements the mutant phenotype of MG5128. (A) Overview of SNEV and Prp19 constructs tested in this study. Length indicates the number of amino acids (aa), MW is the molecular weight (kd), (+) indicates ability to grow at the indicated temperature, to sporulate or to perform forward mutability. (−) no growth, (n.d.) not determined. The star in construct d indicates the point mutation. (B) All chimeric constructs introduced into the mutant yeast strain are translated into protein. Western blots after SDS–PAGE of cell-lysates of MG5128 transformed with pYPGE15, containing various chimeric constructs, which were fused to a His6-tag. Detection was performed using anti-TetraHis mouse antibody as primary and peroxidase-conjugated goat-anti-mouse antibody as secondary antibody. Calculated molecular masses [kDa] are indicated. The small bands in lanes 4–6 and 11–14 represent degradation products resulting from the lysis process. (C) Growth at the permissive (30°C) and the non-permissive (37°C) temperature of MG5128, transformed with the various constructs was analysed. All clones showed growth at 30°C, whereas at 37°C only transformants containing wtPrp19p, SNEV[1–66]Prp19p[68–503], Prp19p[68–503] or SNEV[1–91]Prp19p[98–503] were able to form colonies. Sector 1: wt Prp19p (positive control), 2: empty vector control, 3: SNEV, 4: SNEV[1–66]Prp19p[68–503], 5: Prp19p[68–503], 6: SNEV[1–91]Prp19p[98–503], 7: Prp19p[97–503], 8: SNEV[1–205]Prp19p[208–503]. (D) Ability of the various constructs to sporulate on KAC agar plates. MG5128 transformed with the various constructs was incubated at 30°C for one week, vegetative and sporulated cells were counted. Sporulation was calculated as the ratio of sporulated versus vegetative cells. Mean and standard deviation (SD) were calculated from at least three independent experiments. The total number of cells counted is indicated. (E) The human–yeast chimeric protein SNEV[1–66]Prp19[68–503] restores wild-type phenotype regarding UV sensitivity and forward mutability. Survival of the various MG5128 transformants after UV-irradiation (left panel) (266 nm) and ability of the various chimeric constructs to restore wild-type like forward mutability by growth on canavanine containing media after UV-irradiation (right panel). Number of mutants has been calculated as mutants per UV radiation surviving cells.
Figure 3
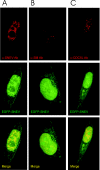
SNEV associates in vivo with Sm proteins as well as with the splicing factor CDC5L, and fusion of SNEV to EGFP does not alter the cellular localization of SNEV. HeLa cells were grown, transfected with an EGFP–SNEV fusion construct, fixed and stained by indirect immunofluorescence. The images shown in the panels are representative optical sections from the respective deconvolved datasets. (A) GFP–SNEV expressing HeLa cells stained with anti-SNEV antibody 867. Red represents SNEV indirect staining, and green indicates EGFP–SNEV expression. (B) Cells were stained with the the anti-Sm protein monoclonal antibody Y12 (57). Red indicates Y12 staining as above, while green represents EGFP–SNEV expression. (C) Cells stained with anti-CDC5L antibodies. Red represents anti-CDC5L staining, while green shows EGFP–SNEV expression. In all the panels, yellow indicates co-localization of the two proteins. Note that the use of the fluorescently labelled protein results in a stronger signal from the fusion protein. This shows up as a small fraction of the label that does not co-localize.
Figure 4
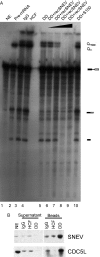
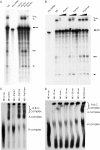
Depletion of SNEV decreases the splicing efficiency of nuclear extracts. (A) After immunodepletion of SNEV from nuclear extracts (NE) treated with 1 M NaCl in order to disrupt the CDC5L complex the extracts were tested for splicing efficiency. Lane 1 shows the unspliced adenoviral pre-mRNA. A decrease of the second catalytical step of the reaction was observed in the SNEV depleted extracts when compared to depleted controls, where irrelevant antibodies were used for depletion (lanes 3 and 4) or where untreated NE was used for in vitro splicing (lane 2). Splicing efficiency is restored, when S100 cytoplasmic extract enriched for splicing factors is added to the SNEV depleted NE (lane 10). However, add back of increasing amounts of recombinant SNEV results in concentration dependent inhibition of the first step of the reaction (lanes 6–9). (B) The antibody used to immunodeplete the high salt nuclear extracts recognizes a distinct band at the expected molecular weight of 56 kDa in nuclear extract (lane NE). While neither IgG nor an irrelevant antibody directed against HCF reduce the amount of SNEV in the nuclear extracts used in the splicing assay after immunodepletion, a marked decrease in SNEV was observed after depletion with α-SNEV antibody. This is consistent with the larger amounts that are eluted from the α-SNEV antibody coupled beads. Although 1 M NaCl was used to disrupt the CDC5L associated complex, CDC5L was co-precipitated together with SNEV. Supernatant: Nuclear extracts after immunodepletion. Beads: Precipitates eluted from the beads. NE (nuclear extract), Igg: IgG coupled beads, HCF: α-HCF antibody coupled beads, DD: α-SNEV antibody coupled beads.
Figure 5
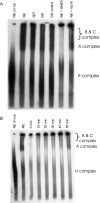
SNEV inhibits spliceosome formation, but does not disrupt already formed complexes. (A) Formation of the splicing complexes was observed by separation on native gels. Lane 1: nuclear extract (NE) on ice as negative control, lane 2: nuclear extract, lane 3: NE after treating with pre-immune IgG coupled beads, lane 4: NE after SNEV depletion, lane 5: NE after treating with anti-SNEV coupled beads pre-incubated with recombinant His6–SNEV, lane 6: NE after addition of recombinant His6-SNEV, lane 7: NE after addition of the same amount of recombinant His6-VP16 proteins as control. Immunodepletion of SNEV only slightly decreases formation of the spliceosome, while after addition of recombinant SNEV no splicing complexes can be detected. (B) Already formed complexes are not disrupted by addition of SNEV in a time course experiment. Lane 1: NE on ice, lane 2: NE, lane 3: NE with SNEV added before starting the reaction, lane 4: SNEV was added 10 min after starting of the reaction, lane 5: 20 min; lane 6: 30 min, lane 7: 40 min, lane 8: 50 min after starting the reaction.
Figure 6
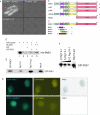
SNEV forms homo-oligomeres that are dependent on the amino acids 56–92 of SNEV. (A) SNEV interacts with SNEV in a yeast two-hybrid assay when GAL4-AD-SNEV and GAL4-BD-SNEV are co-transformed into the yeast reporter strain AH109 (AD-SNEV + BD-SNEV). Neither co-transformation of SNEV fusion to the GAL4 activating domain (AD-SNEV + BD) nor to the DNA-binding domain (BD-SNEV + AD) together with the respective empty second plasmid allowed formation of yeast colonies on high stringency drop-out media plates. (B) Domain mapping of the self interacting amino acids was performed using the Yeast two-hybrid system. Various truncated SNEV constructs were used as prey. Growth of double transformants on high stringency media plates was observed only for variants containing amino acids 68–90, which we therefore termed self interaction domain (SID). (C) In order to confirm that the SID is sufficient for homo-oligomerization, four small overlapping peptides were cloned and tested by Y2H. Co-transformation with SNEV resulted in growth of yeast double transformants on high stringency media plates only with the region 56–92 and 56–121 (red), while neither 66–103 nor 76–121 (green) allowed yeast colony formation. U-box: domain necessary for ubiquitin E3 ligase activity of SNEV; SID: Self Interaction domain; LCR: low complexity region; GL2: globular domain 2, WD40: domain containing 7 WD40 repeats; numbers indicate the amino acid positions, ‘+’ indicates colony formation on high stringency drop out media, ‘−’ indicates no colony formation. (D) The SNEV self-interaction was confirmed by GST pull-down experiments using affinity purified His6–SNEV and GST–SNEV. While no His6–SNEV was detectable in the controls using either GST or beads alone (lanes 2 and 3), GST–SNEV results in co-precipitation of His6–SNEV (lanes 4 and 5). (E) Addition of Prp19-1 peptide to GST–SNEV reduces the amount of precipitated SNEV (Prp19-1), whereas addition of the scrambled control peptide (Prp19-scr) does not. This amount is comparable to the one without addition of peptides (GST–SNEV) as well as to the amount of GST–SNEV added to the beads (Input). (F) Increasing amounts of the Prp19-1 peptide (0, 5 and 10 nM) decrease the amount of precipitated GST–SNEV (G) Additional confirmation of SNEV self interaction derives from FRET analysis. Upper panel: Co-transfection of ECFP–SNEV and EYFP–SNEV into HeLa cells resulted. Pictures were taken using CFP-filter (ECFP–SNEV) displayed in cyan, YFP-filter (EYFP–SNEV). FRET represents the calculated net FRET signal. Lower panel: negative control using ECFP–SNEVΔ98 lacking the interaction domain.
Figure 7
A peptide that spans the SNEV self interaction site inhibits the splicing reaction by interfering with spliceosome stability. (A) Overlapping peptides that span the self interaction site of SNEV were synthesized and added to standard splicing reactions. While nuclear extract (NE) as positive control shows efficient splicing of an adenoviral pre-mRNA, the peptides Prp19-1 and Prp19-4 show inhibition of the splicing reaction, while Prp19-2 and Prp19-3 do not affect formation of lariat and of joined exons. (B) Increasing amounts (7, 14 and 21 nmol) of the two effective peptides were added to the splicing reaction, showing higher potency for Prp19-1 than for Prp19-4. As negative control the same amounts of Prp19-scr, the scrambled version of Prp19-1 was used. (C) Splicing complexes have been separated on a native gel. Addition of Prp19-1 (NE + Prp19-1) peptide results in inhibition of formation of C, B and A splicing complexes similar to incubation of the splicing reaction on ice (NE on ice), while Prp19-4 (NE + Prp19-4) as well as Prp19-scr (NE + Prp19-scr) similar to the positive control (NE) do not show any reduction in spliceosome assembly. (D) 21 nmol Prp19-1 were added to splicing reactions at different time points: either pre-incubation for 10 min (−10 min), at the start of the reaction (0 min), or at 10 min, 25 min and 50 min after starting the splicing reaction. After 55 min all reactions were loaded on to native gels. Neither A, B nor C complexes can be detected independent from the time point of peptide addition, while the scrambled peptide does not interfere with spliceosome formation.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - "I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.
Huang Y, Trollor JN, Foley KR, Arnold SRC. Huang Y, et al. Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. PMID: 28752910 Free PMC article. Updated. Review.
Cited by
- Pre-mRNA processing factors meet the DNA damage response.
Montecucco A, Biamonti G. Montecucco A, et al. Front Genet. 2013 Jun 6;4:102. doi: 10.3389/fgene.2013.00102. eCollection 2013. Front Genet. 2013. PMID: 23761808 Free PMC article. - A proteomic comparison of immature and mature mouse gonadotrophs reveals novel differentially expressed nuclear proteins that regulate gonadotropin gene transcription and RNA splicing.
Feng J, Lawson MA, Melamed P. Feng J, et al. Biol Reprod. 2008 Sep;79(3):546-61. doi: 10.1095/biolreprod.108.068106. Epub 2008 May 14. Biol Reprod. 2008. PMID: 18480465 Free PMC article. - Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity.
Monaghan J, Xu F, Gao M, Zhao Q, Palma K, Long C, Chen S, Zhang Y, Li X. Monaghan J, et al. PLoS Pathog. 2009 Jul;5(7):e1000526. doi: 10.1371/journal.ppat.1000526. Epub 2009 Jul 24. PLoS Pathog. 2009. PMID: 19629177 Free PMC article. - Ubiquitous overexpression of the DNA repair factor dPrp19 reduces DNA damage and extends Drosophila life span.
Garschall K, Dellago H, Gáliková M, Schosserer M, Flatt T, Grillari J. Garschall K, et al. NPJ Aging Mech Dis. 2017 Mar 15;3:5. doi: 10.1038/s41514-017-0005-z. eCollection 2017. NPJ Aging Mech Dis. 2017. PMID: 28649423 Free PMC article. - SNEVhPrp19/hPso4 Regulates Adipogenesis of Human Adipose Stromal Cells.
Khan A, Dellago H, Terlecki-Zaniewicz L, Karbiener M, Weilner S, Hildner F, Steininger V, Gabriel C, Mück C, Jansen-Dürr P, Hacobian A, Scheideler M, Grillari-Voglauer R, Schosserer M, Grillari J. Khan A, et al. Stem Cell Reports. 2017 Jan 10;8(1):21-29. doi: 10.1016/j.stemcr.2016.12.001. Epub 2016 Dec 29. Stem Cell Reports. 2017. PMID: 28041875 Free PMC article.
References
- Grillari J., Hohenwarter O., Grabherr R.M., Katinger H. Subtractive hybridization of mRNA from early passage and senescent endothelial cells. Exp. Gerontol. 2000;35:187–197. - PubMed
- Voglauer R., Chang M., Wieser M., Dampier B., Baumann K., Schreiber M., Katinger H., Grillari J. Overexpression of SNEV extends the replicative life span of human endothelial cells. Exp. Cell Res. in press. - PubMed
- Chen H.R., Jan S.P., Tsao T.Y., Sheu Y.J., Banroques J., Cheng S.C. SNT309p, a component of the Prp19p-associated comples that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 1998;18:2196–2204. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous