PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample - PubMed (original) (raw)
PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample
Silvia G Acinas et al. Appl Environ Microbiol. 2005 Dec.
Abstract
The contribution of PCR artifacts to 16S rRNA gene sequence diversity from a complex bacterioplankton sample was estimated. Taq DNA polymerase errors were found to be the dominant sequence artifact but could be constrained by clustering the sequences into 99% sequence similarity groups. Other artifacts (chimeras and heteroduplex molecules) were significantly reduced by employing modified amplification protocols. Surprisingly, no skew in sequence types was detected in the two libraries constructed from PCR products amplified for different numbers of cycles. Recommendations for modification of amplification protocols and for reporting diversity estimates at 99% sequence similarity as a standard are given.
Figures
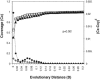
FIG. 1.
LIBSHUFF coverage curve comparison for the modified and standard libraries. Shown are the homologous coverage curve (Cx) of the modified library (squares), the heterologous coverage curve (Cxy) of the modified library versus the standard library (triangles), and the value of (Cx-Cxy)2 (crosses) for the samples at each D value. D is equal to the Jukes-Cantor evolutionary distance determined using PAUP software. Filled circles indicate the P value of 0.05 for (Cx-Cxy)2 for the randomized samples.
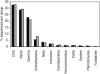
FIG. 2.
Relative frequency distribution of major phylogenetic groups detected among the environmental 16S rRNA sequences from the standard (gray bars) and modified (black bars) libraries, respectively.
References
- Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources