Selective sweep mapping of genes with large phenotypic effects - PubMed (original) (raw)
Selective sweep mapping of genes with large phenotypic effects
John P Pollinger et al. Genome Res. 2005 Dec.
Abstract
Many domestic dog breeds have originated through fixation of discrete mutations by intense artificial selection. As a result of this process, markers in the proximity of genes influencing breed-defining traits will have reduced variation (a selective sweep) and will show divergence in allele frequency. Consequently, low-resolution genomic scans can potentially be used to identify regions containing genes that have a major influence on breed-defining traits. We model the process of breed formation and show that the probability of two or three adjacent marker loci showing a spurious signal of selection within at least one breed (i.e., Type I error or false-positive rate) is low if highly variable and moderately spaced markers are utilized. We also use simulations with selection to demonstrate that even a moderately spaced set of highly polymorphic markers (e.g., one every 0.8 cM) has high power to detect regions targeted by strong artificial selection in dogs. Further, we show that a gene responsible for black coat color in the Large Munsterlander has a 40-Mb region surrounding the gene that is very low in heterozygosity for microsatellite markers. Similarly, we survey 302 microsatellite markers in the Dachshund and find three linked monomorphic microsatellite markers all within a 10-Mb region on chromosome 3. This region contains the FGFR3 gene, which is responsible for achondroplasia in humans, but not in dogs. Consequently, our results suggest that the causative mutation is a gene or regulatory region closely linked to FGFR3.
Figures
Figure 1.
Effects of time since breed formation on FST and observed marker heterozygosity. Expected levels of marker heterozygosity under the infinite-alleles model (E(H) = θ/(1 + θ)); (gray lines). Observed heterozygosity after conditioning on markers showing variability among breeds (black lines).
Figure 2.
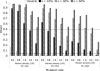
Effect of heterozygosity on Type I error of all statistics considered in this study across 500,000 simulated replicate data sets. Simulations simultaneously varied time since breed formation, number of breeds compared, number of chromosomes sampled per breed, extent of bottleneck, duration of bottleneck, marker variability and density, and size of ancestral population.
Figure 3.
Simulation results summarizing the effect of selection on the average heterozygosity and FST among 200 replicate data sets per parameter combination (selection ranging from 10%-50% for marker density of 0.8 cM and 3.2 cM spacing between markers).
Figure 4.
Summary of the power of the homozygosity statistics with varying values of recombination, selection, and mutation.
Figure 5.
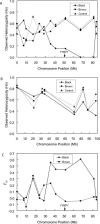
Microsatellite marker heterozygosity (A,B) and FST (C) values for the Large Munsterlander (Black), German Longhair (Brown), and control breeds. (A) Heterozygosity for markers on chromosome 11, the location of the TYRP1 gene, (B) heterozygosity for markers on control chromosome 5, (C) FST values for markers on chromosome 11.
Figure 6.
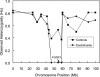
Heterozygosity for markers on chromosome 3 for Dachshunds and control breeds. A low heterozygosity sweep region unique to Dachshunds and including the FGFR3 gene is found.
Figure 7.
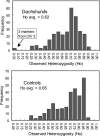
Heterozygosity frequency distribution for 302 microsatellite markers in Dachshunds (top) and control breeds (bottom).
Similar articles
- So many doggone traits: mapping genetics of multiple phenotypes in the domestic dog.
Rimbault M, Ostrander EA. Rimbault M, et al. Hum Mol Genet. 2012 Oct 15;21(R1):R52-7. doi: 10.1093/hmg/dds323. Epub 2012 Aug 9. Hum Mol Genet. 2012. PMID: 22878052 Free PMC article. Review. - Localization of canine brachycephaly using an across breed mapping approach.
Bannasch D, Young A, Myers J, Truvé K, Dickinson P, Gregg J, Davis R, Bongcam-Rudloff E, Webster MT, Lindblad-Toh K, Pedersen N. Bannasch D, et al. PLoS One. 2010 Mar 10;5(3):e9632. doi: 10.1371/journal.pone.0009632. PLoS One. 2010. PMID: 20224736 Free PMC article. - Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping.
Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppälä EH, Hansen MS, Lawley CT, Karlsson EK; LUPA Consortium; Bannasch D, Vilà C, Lohi H, Galibert F, Fredholm M, Häggström J, Hedhammar A, André C, Lindblad-Toh K, Hitte C, Webster MT. Vaysse A, et al. PLoS Genet. 2011 Oct;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022279 Free PMC article. - A selective sweep of >8 Mb on chromosome 26 in the Boxer genome.
Quilez J, Short AD, Martínez V, Kennedy LJ, Ollier W, Sanchez A, Altet L, Francino O. Quilez J, et al. BMC Genomics. 2011 Jul 1;12:339. doi: 10.1186/1471-2164-12-339. BMC Genomics. 2011. PMID: 21722374 Free PMC article. - The canine genome.
Ostrander EA, Wayne RK. Ostrander EA, et al. Genome Res. 2005 Dec;15(12):1706-16. doi: 10.1101/gr.3736605. Genome Res. 2005. PMID: 16339369 Review.
Cited by
- Genetic hitchhiking in a subdivided population of Mytilus edulis.
Faure MF, David P, Bonhomme F, Bierne N. Faure MF, et al. BMC Evol Biol. 2008 May 30;8:164. doi: 10.1186/1471-2148-8-164. BMC Evol Biol. 2008. PMID: 18513403 Free PMC article. - Detecting directional selection in the presence of recent admixture in African-Americans.
Lohmueller KE, Bustamante CD, Clark AG. Lohmueller KE, et al. Genetics. 2011 Mar;187(3):823-35. doi: 10.1534/genetics.110.122739. Epub 2010 Dec 31. Genetics. 2011. PMID: 21196524 Free PMC article. - A genome scan for selection signatures comparing farmed Atlantic salmon with two wild populations: Testing colocalization among outlier markers, candidate genes, and quantitative trait loci for production traits.
Liu L, Ang KP, Elliott JA, Kent MP, Lien S, MacDonald D, Boulding EG. Liu L, et al. Evol Appl. 2016 Dec 29;10(3):276-296. doi: 10.1111/eva.12450. eCollection 2017 Mar. Evol Appl. 2016. PMID: 28250812 Free PMC article. - Screen for Footprints of Selection during Domestication/Captive Breeding of Atlantic Salmon.
Vasemägi A, Nilsson J, McGinnity P, Cross T, O'Reilly P, Glebe B, Peng B, Berg PR, Primmer CR. Vasemägi A, et al. Comp Funct Genomics. 2012;2012:628204. doi: 10.1155/2012/628204. Epub 2012 Dec 27. Comp Funct Genomics. 2012. PMID: 23326209 Free PMC article. - So many doggone traits: mapping genetics of multiple phenotypes in the domestic dog.
Rimbault M, Ostrander EA. Rimbault M, et al. Hum Mol Genet. 2012 Oct 15;21(R1):R52-7. doi: 10.1093/hmg/dds323. Epub 2012 Aug 9. Hum Mol Genet. 2012. PMID: 22878052 Free PMC article. Review.
References
- American Kennel Club. 1997. The complete dog book; the histories and standards of breeds admitted to AKC registration, and the feeding, training, care, breeding, and health of pure-bred dogs,19th ed. Doubleday, Garden City, NY.
- Ash, E.C. 1927. Dogs: Their history and development. E. Benn Limited, London.
Web site references
- http://www2.unil.ch/popgen/softwares/fstat.htm; Fstat Version 2.9.3.2.
- http://research.nhgri.nih.gov/dog_genome/guyon2003/; The 1-Mb resolution radiation hybrid map of the canine genome (Guyon et al. 2003). - PMC - PubMed
- http://www.eeb.ucla.edu/dogmarkerset/; The 302 microsatellite marker set used in this study.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous