An antiviral role for the RNA interference machinery in Caenorhabditis elegans - PubMed (original) (raw)
An antiviral role for the RNA interference machinery in Caenorhabditis elegans
Daniel H Schott et al. Proc Natl Acad Sci U S A. 2005.
Abstract
RNA interference (RNAi) is a sequence-specific gene-silencing mechanism triggered by exogenous dsRNA. In plants an RNAi-like mechanism defends against viruses, but the hypothesis that animals possess a similar natural antiviral mechanism related to RNAi remains relatively untested. To test whether genes needed for RNAi defend animal cells against virus infection, we infected wild-type and RNAi-defective cells of the nematode C. elegans with vesicular stomatitis virus engineered to encode a GFP fusion protein. We show that upon infection, cells lacking components of the RNAi apparatus produce more GFP and infective particles than wild-type cells. Furthermore, we show that mutant cells with enhanced RNAi produce less GFP. Our observation that multiple genes required for RNAi are also required for resistance to vesicular stomatitis virus suggests that the RNAi machinery functions in resistance to viruses in nature.
Figures
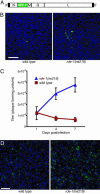
Fig. 1.
VSV infection of wild-type and rde-1 mutant C. elegans cells. (A) Schematic of the genome structure of the recombinant VSV used in this study, encoding a GFP–P fusion protein. (B) C. elegans cells 7 days after infection at moi = 0.003 (15). Green/yellow, GFP fluorescence; blue, Hoechst 33342 staining for DNA. (Scale bar, 100 μm.) (C) Time course of virus titers capable of infecting mammalian cells, in medium after infection of wild type and rde-1(ne219) cells at moi = 3. Error bars represent SEM; n = 3 cell-culture wells. Titer in medium over rde-1(ne219) cells at day 7 is different from titer over wild-type cells at day 7 (P = 0.03 in two-tailed t test). (D) C. elegans cells 7 days after infection at moi = 3. Green, GFP fluorescence; blue, Hoechst 33342 staining for DNA. (Scale bar, 50 μm.)
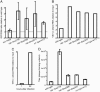
Fig. 2.
VSV replication in infected wild-type and RNAi-defective C. elegans cells. (A) GFP levels in RNAi-defective mutants relative to wild-type 7 days after infection at moi = 3. Values are ratios of GFP fluorescence to Hoechst 33342 fluorescence, relative to wild type. Error bars represent the 95% confidence interval in t distribution based on four to eight worm embryonic cell isolations. Different from wild type in two-tailed t tests: rde-1(ne219), P = 7 × 10–4; rde-3(ne298), P = 0.03; rde-4(ne301), P = 0.02; rrf-1(pk1417), P = 2 × 10–4; sid-1(qt9), P = 0.07. Different from sid-1(qt9) in two-tailed t tests: rde-1(ne219), P = 9 × 10–4; rde-3(ne298), P = 0.04; rde-4(ne301), P = 0.02; rrf-1(pk1417), P = 1 × 10–5. (B) Quantities of VSV (–)-strand RNA in cells 7 days after infection at moi = 3, relative to virus used to infect cells. (C) Quantities of VSV (–)-strand RNA in rde-1(ne219) cells 1 h and 52 h after infection at moi = 3, relative to virus used to infect cells. (D) Virus titers capable of infecting mammalian cells, in medium 7 days after infection of wild-type and mutant cells at moi = 3. Error bars represent SEM; n = 4 cell culture wells. Different from wild type in two-tailed t tests: rde-1(ne219), P = 1 × 10–3; rde-3(ne298), P = 4 × 10–3; rde-4(ne301), P = 7 × 10–6; rrf-1(pk1417), P = 2 × 10–3.
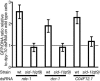
Fig. 3.
GFP levels in dsRNA-treated wild-type and sid-1(qt9) cell cultures relative to wild type (wt) treated with irrelevant (dpy-10) dsRNA 7 days after infection at moi = 3. Values are ratios of GFP fluorescence to Hoechst 33342 fluorescence, relative to wild type. Error bars represent the 95% confidence interval in t distribution based on samples within one experiment, but data are representative of at least two experiments. Different from dpy-10 dsRNA-treated wild type in two-tailed t tests: rde-1 dsRNA-treated wild type, P = 2 × 10–12; dcr-1 dsRNA-treated wild type, P = 2 × 10–7; C04F12.1 dsRNA-treated wild type, P = 7 × 10–12. Different from corresponding sid-1(qt9) cells in two-tailed t tests: rde-1 dsRNA-treated wild type, P = 7 × 10–13; dcr-1 dsRNA-treated wild type, P = 1 × 10–8; C04F12.1 dsRNA-treated wild type, P = 7 × 10–10.
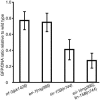
Fig. 4.
GFP levels in RNAi enhanced mutants relative to wild type 7 days after infection at moi = 6. Values are ratios of GFP fluorescence to Hoechst 33342 fluorescence, relative to wild type. Error bars represent the 95% confidence interval in t distribution based on samples within one experiment, but data are representative of at least two experiments. Different from wild type in two-tailed t tests: rrf-3(pk1426), P = 6 × 10–4; eri-1(mg366), P = 3 × 10–4; lin-15B(n744), P = 2 × 10–8; eri-1(mg366);lin-15B(n744), P = 2 × 10–8.
Similar articles
- Transgene-Assisted Genetic Screen Identifies rsd-6 and Novel Genes as Key Components of Antiviral RNA Interference in Caenorhabditis elegans.
Long T, Meng F, Lu R. Long T, et al. J Virol. 2018 Aug 16;92(17):e00416-18. doi: 10.1128/JVI.00416-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950414 Free PMC article. - An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans.
Lu R, Yigit E, Li WX, Ding SW. Lu R, et al. PLoS Pathog. 2009 Feb;5(2):e1000286. doi: 10.1371/journal.ppat.1000286. Epub 2009 Feb 6. PLoS Pathog. 2009. PMID: 19197349 Free PMC article. - Antiviral RNA Interference against Orsay Virus Is neither Systemic nor Transgenerational in Caenorhabditis elegans.
Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. Ashe A, et al. J Virol. 2015 Dec;89(23):12035-46. doi: 10.1128/JVI.03664-14. Epub 2015 Sep 23. J Virol. 2015. PMID: 26401037 Free PMC article. - Small RNA-mediated gene silencing pathways in C. elegans.
Fischer SE. Fischer SE. Int J Biochem Cell Biol. 2010 Aug;42(8):1306-15. doi: 10.1016/j.biocel.2010.03.006. Epub 2010 Mar 19. Int J Biochem Cell Biol. 2010. PMID: 20227516 Review. - Environmental RNA interference.
Whangbo JS, Hunter CP. Whangbo JS, et al. Trends Genet. 2008 Jun;24(6):297-305. doi: 10.1016/j.tig.2008.03.007. Epub 2008 Apr 29. Trends Genet. 2008. PMID: 18450316 Review.
Cited by
- Guest list or black list: heritable small RNAs as immunogenic memories.
Rechavi O. Rechavi O. Trends Cell Biol. 2014 Apr;24(4):212-20. doi: 10.1016/j.tcb.2013.10.003. Epub 2013 Nov 11. Trends Cell Biol. 2014. PMID: 24231398 Free PMC article. Review. - Slicing and dicing viruses: antiviral RNA interference in mammals.
Maillard PV, van der Veen AG, Poirier EZ, Reis e Sousa C. Maillard PV, et al. EMBO J. 2019 Apr 15;38(8):e100941. doi: 10.15252/embj.2018100941. Epub 2019 Mar 14. EMBO J. 2019. PMID: 30872283 Free PMC article. Review. - RNA-based viral immunity initiated by the Dicer family of host immune receptors.
Aliyari R, Ding SW. Aliyari R, et al. Immunol Rev. 2009 Jan;227(1):176-88. doi: 10.1111/j.1600-065X.2008.00722.x. Immunol Rev. 2009. PMID: 19120484 Free PMC article. Review. - Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression.
Cureton DK, Burdeinick-Kerr R, Whelan SP. Cureton DK, et al. J Virol. 2012 Jan;86(2):655-66. doi: 10.1128/JVI.05810-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072764 Free PMC article. - Aedes aegypti uses RNA interference in defense against Sindbis virus infection.
Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Campbell CL, et al. BMC Microbiol. 2008 Mar 17;8:47. doi: 10.1186/1471-2180-8-47. BMC Microbiol. 2008. PMID: 18366655 Free PMC article.
References
- Meister, G. & Tuschl, T. (2004) Nature 431, 343–349. - PubMed
- Mello, C. C. & Conte, D., Jr. (2004) Nature 431, 338–342. - PubMed
- Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99, 123–132. - PubMed
- Kim, J. K., Gabel, H. W., Kamath, R. S., Tewari, M., Pasquinelli, A., Rual, J. F., Kennedy, S., Dybbs, M., Bertin, N., Kaplan, J. M., et al. (2005) Science 308, 1164–1167. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM069891-04/GM/NIGMS NIH HHS/United States
- R01 AI059371-01A1/AI/NIAID NIH HHS/United States
- R01 GM069891/GM/NIGMS NIH HHS/United States
- R37 AI059371/AI/NIAID NIH HHS/United States
- AI059371/AI/NIAID NIH HHS/United States
- R01 AI059371/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources