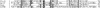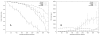Kalign--an accurate and fast multiple sequence alignment algorithm - PubMed (original) (raw)
Kalign--an accurate and fast multiple sequence alignment algorithm
Timo Lassmann et al. BMC Bioinformatics. 2005.
Abstract
Background: The alignment of multiple protein sequences is a fundamental step in the analysis of biological data. It has traditionally been applied to analyzing protein families for conserved motifs, phylogeny, structural properties, and to improve sensitivity in homology searching. The availability of complete genome sequences has increased the demands on multiple sequence alignment (MSA) programs. Current MSA methods suffer from being either too inaccurate or too computationally expensive to be applied effectively in large-scale comparative genomics.
Results: We developed Kalign, a method employing the Wu-Manber string-matching algorithm, to improve both the accuracy and speed of multiple sequence alignment. We compared the speed and accuracy of Kalign to other popular methods using Balibase, Prefab, and a new large test set. Kalign was as accurate as the best other methods on small alignments, but significantly more accurate when aligning large and distantly related sets of sequences. In our comparisons, Kalign was about 10 times faster than ClustalW and, depending on the alignment size, up to 50 times faster than popular iterative methods.
Conclusion: Kalign is a fast and robust alignment method. It is especially well suited for the increasingly important task of aligning large numbers of sequences.
Figures
Figure 1
Balibase 2.01 reference alignment 1tvxA ref1 viewed by Belvu [37], showing conservation by "average similarity by BLOSUM62".
Figure 2
Analysis of the contribution to alignment accuracy made by different algorithmic variants. Kalign-default uses Wu-Manber approximate string matching, while Kalign-ktuple, Mafft-fast, Muscle-fast, and ClustalW-quicktree use exact k-tuple matching. The default Kalign Wu-Manber based algorithm becomes more accurate than other methods at high evolutionary distances. The alignments consisted of 50 simulated sequences.
Figure 3
A 2D plot indicating in which situations different methods perform better on the large testset. The accuracy of the most accurate versions of Kalign, Muscle, and Mafft was measured for each combination of average evolutionary distance (in PAM units) and number of sequences. The cells were colored according to the most accurate program as: Kalign:red; Muscle:blue; Mafft:yellow. If there was a tie between two or more methods the cell is black. In (a) it is enough to win by the smallest margin, whereas in (b) the program must win by a margin of 2%. Up to 200 PAM no program stands out as a clear winner while above this distance Kalign dominates.
Figure 4
Plots of the accuracy (a) and speed (b) achieved by of Kalign, Mafft (FFTNSI), Muscle, and ClustalW on the large testset with increasing average evolutionary distance. The number of sequences (300) and the average sequence length (500 residues) are kept constant.
Figure 5
Plots of the accuracy (a) and speed (b) achieved by of Kalign, Mafft (FFTNSI), Muscle, and ClustalW on the large testset with increasing number of sequences. The evolutionary distance (300 PAM) and the average sequence length (500 residues) are kept constant.
Similar articles
- transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences.
Bininda-Emonds OR. Bininda-Emonds OR. BMC Bioinformatics. 2005 Jun 22;6:156. doi: 10.1186/1471-2105-6-156. BMC Bioinformatics. 2005. PMID: 15969769 Free PMC article. - Grammar-based distance in progressive multiple sequence alignment.
Russell DJ, Otu HH, Sayood K. Russell DJ, et al. BMC Bioinformatics. 2008 Jul 10;9:306. doi: 10.1186/1471-2105-9-306. BMC Bioinformatics. 2008. PMID: 18616828 Free PMC article. - TM-Aligner: Multiple sequence alignment tool for transmembrane proteins with reduced time and improved accuracy.
Bhat B, Ganai NA, Andrabi SM, Shah RA, Singh A. Bhat B, et al. Sci Rep. 2017 Oct 2;7(1):12543. doi: 10.1038/s41598-017-13083-y. Sci Rep. 2017. PMID: 28970546 Free PMC article. - Multiple sequence alignment.
Edgar RC, Batzoglou S. Edgar RC, et al. Curr Opin Struct Biol. 2006 Jun;16(3):368-73. doi: 10.1016/j.sbi.2006.04.004. Epub 2006 May 5. Curr Opin Struct Biol. 2006. PMID: 16679011 Review. - Differences between pair-wise and multi-sequence alignment methods affect vertebrate genome comparisons.
Margulies EH, Chen CW, Green ED. Margulies EH, et al. Trends Genet. 2006 Apr;22(4):187-93. doi: 10.1016/j.tig.2006.02.005. Epub 2006 Feb 24. Trends Genet. 2006. PMID: 16499991 Review.
Cited by
- Correction: genomic comparison of 93 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity.
Grose JH, Jensen GL, Burnett SH, Breakwell DP. Grose JH, et al. BMC Genomics. 2014 Dec 29;15(1):1184. doi: 10.1186/1471-2164-15-1184. BMC Genomics. 2014. PMID: 25547158 Free PMC article. - A spruce gene map infers ancient plant genome reshuffling and subsequent slow evolution in the gymnosperm lineage leading to extant conifers.
Pavy N, Pelgas B, Laroche J, Rigault P, Isabel N, Bousquet J. Pavy N, et al. BMC Biol. 2012 Oct 26;10:84. doi: 10.1186/1741-7007-10-84. BMC Biol. 2012. PMID: 23102090 Free PMC article. - Dynamic Transcriptome Sequencing of Bovine Alphaherpesvirus Type 1 and Host Cells Carried Out by a Multi-Technique Approach.
Tombácz D, Moldován N, Torma G, Nagy T, Hornyák Á, Csabai Z, Gulyás G, Boldogkői M, Jefferson VA, Zádori Z, Meyer F, Boldogkői Z. Tombácz D, et al. Front Genet. 2021 Apr 7;12:619056. doi: 10.3389/fgene.2021.619056. eCollection 2021. Front Genet. 2021. PMID: 33897757 Free PMC article. No abstract available. - MsPAC: a tool for haplotype-phased structural variant detection.
Rodriguez OL, Ritz A, Sharp AJ, Bashir A. Rodriguez OL, et al. Bioinformatics. 2020 Feb 1;36(3):922-924. doi: 10.1093/bioinformatics/btz618. Bioinformatics. 2020. PMID: 31397844 Free PMC article. - Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin.
Almén MS, Nordström KJ, Fredriksson R, Schiöth HB. Almén MS, et al. BMC Biol. 2009 Aug 13;7:50. doi: 10.1186/1741-7007-7-50. BMC Biol. 2009. PMID: 19678920 Free PMC article.
References
- Notredame C. Recent progress in multiple sequence alignment: a survey. Pharmacogenomics. 2002;3:131–144. - PubMed
- Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166.
- Sjolander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–179. - PubMed
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources