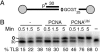Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1 - PubMed (original) (raw)
Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1
Parie Garg et al. Proc Natl Acad Sci U S A. 2005.
Abstract
In response to DNA damage, the Rad6/Rad18 ubiquitin-conjugating complex monoubiquitinates the replication clamp proliferating cell nuclear antigen (PCNA) at Lys-164. Although ubiquitination of PCNA is recognized as an essential step in initiating postreplication repair, the mechanistic relevance of this modification has remained elusive. Here, we describe a robust in vitro system that ubiquitinates yeast PCNA specifically on Lys-164. Significantly, only those PCNA clamps that are appropriately loaded around effector DNA by its loader, replication factor C, are ubiquitinated. This observation suggests that, in vitro, only PCNA present at stalled replication forks is ubiquitinated. Ubiquitinated PCNA displays the same replicative functions as unmodified PCNA. These functions include loading onto DNA by replication factor C, as well as Okazaki fragment synthesis and maturation by the PCNA-coordinated actions of DNA polymerase delta, the flap endonuclease FEN1, and DNA ligase I. However, whereas the activity of DNA polymerase zeta remains unaffected by ubiquitination of PCNA, ubiquitinated PCNA specifically activates two key enzymes in translesion synthesis: DNA polymerase eta, the yeast Xeroderma pigmentosum ortholog, and Rev1, a deoxycytidyl transferase that functions in organizing the mutagenic DNA replication machinery. We propose that ubiquitination of PCNA increases its functionality as a sliding clamp to promote mutagenic DNA replication.
Figures
Fig. 1.
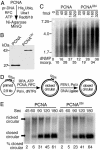
DNA-bound PCNA is ubiquitinated at Lys-164. (A) Scheme of the assay. (B) Factor requirement for efficient PCNA ubiquitination. (C) Human ubiquitin is linked to PCNA. The standard PCNA ubiquitination assay (described in Materials and Methods) contained either the tagged yeast ubiquitin (His6-Ubi, left lane) or human ubiquitin (right lane). (D) Time course of ubiquitination. Assays were carried out on unprimed or DECAprimed ssDNA. (E) Ubiquitination of mutant PCNAs. Standard assays contained unlabeled PCNA or the indicated mutant PCNA. Detection was by Western blot analysis with rabbit antibodies to PCNA.
Fig. 2.
Biochemical properties of PCNAUbi in DNA replication. (A) Purification scheme. Pr-DNA is DECAprimed Bluescript SKII ssDNA. (B) Gel analysis (11% SDS/PAGE) of PCNA and purified PCNAUbi. Staining was with colloidal Coomassie blue. Scanning of the gel showed that ≈3% nonubiquitinated PCNA remained in the purified preparation. (C) Replication of RPA-coated singly primed mp18 ssDNA (60-fmol circles) by Pol δ (120 fmol) and RFC (120 fmol) with increasing PCNA or PCNAUbi was performed essentially as described (38). [α-32P]dTTP was used as radioactive tracer. The PCNA was loaded by RFC for 1 min at 30°C, Pol δ was added, and incubation continued for an additional 90 sec. Products were analyzed on a 1% alkaline agarose gel. (D) Scheme for measuring Okazaki fragment maturation kinetics. (E) Replication and maturation assays of RPA-coated circular ssDNA (100 fmol), primed with an RNA8DNA22 primer, by RFC (200 fmol), PCNA or PCNAUbi (300 fmol), Pol δ (200 fmol), FEN1 (200 fmol), and DNA ligase I (500 fmol) were exactly as described (28). Replication products after the indicated times were separated on a 1% agarose gel in the presence of 0.5 μg/ml ethidium bromide. This method separates covalently closed circular DNA from nicked circular DNA. [α-32P]dTTP was used as radioactive tracer.
Fig. 3.
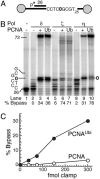
PCNAUbi activates TLS of abasic sites by Pol η.(A) Scheme of substrate V9AP1/C12-4. The gray circles indicate the biotin-streptavidin blocks. The abasic site is shown as 0. (B) Stimulation of bypass by PCNA or PCNAUbi. Standard assays (described in Materials and Methods) were for 5 min with the indicated polymerase and the indicated form of PCNA. The template sequence of interest is shown at left. (C) PCNAUbi stimulates abasic bypass by Pol η. Standard assays with increasing clamp levels were carried out for 60 sec at 30°C. Full-length replication products as the percentage of total primer extension products were quantitated.
Fig. 4.
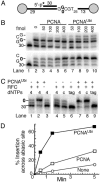
PCNAUbi activates TLS of abasic sites by Rev1. (A) Scheme of substrates V9/C12 and V9AP2/C12. The abasic site is shown as 0.(B) PCNAUbi stimulates Rev1. Standard assays (described in Materials and Methods) contained either the normal template (Upper) or the abasic site template (Lower) and the indicated levels of clamp. The template sequence of interest is shown at left. (C) Control assays for PCNAUbi-mediated Rev1 activity. Either PCNAUbi (lane 1) or RFC (lane 3) was omitted from the standard assay with Rev1 (lane 2). Standard assays without PCNA (lanes 4–6) or with PCNAUbi (lanes 7–9) contained all four dNTPs (4, lanes 4 and 7), only dCTP (c, lanes 5 and 8), or dTTP, dATP, and dGTP (tag, lanes 6 and 9). (D) Time-course assays of PCNAUbi-mediated Rev1 activity at the abasic site. Standard assays were followed with time and insertion opposite the abasic site as percentage of total products was quantitated by using PhosphorImager analysis.
Fig. 5.
PCNAUbi activates Rev1 during “standing start” TLS. (A) Scheme of substrate V9AP1/C12. The abasic site is shown as 0.(B) Time course of PCNAUbi-mediated Rev1 activity at an abasic site. Standard assays were used (described in Materials and Methods), and insertion opposite the abasic site (% TLS) was quantitated.
Similar articles
- The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ.
Tellier-Lebegue C, Dizet E, Ma E, Veaute X, Coïc E, Charbonnier JB, Maloisel L. Tellier-Lebegue C, et al. PLoS Genet. 2017 Dec 27;13(12):e1007119. doi: 10.1371/journal.pgen.1007119. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29281621 Free PMC article. - Rad6/Rad18 Competes with DNA Polymerases η and δ for PCNA Encircling DNA.
Li M, Larsen L, Hedglin M. Li M, et al. Biochemistry. 2020 Feb 4;59(4):407-416. doi: 10.1021/acs.biochem.9b00938. Epub 2020 Jan 10. Biochemistry. 2020. PMID: 31887036 - Ubiquitin-dependent regulation of translesion polymerases.
Chun AC, Jin DY. Chun AC, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review. - Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.
Kanao R, Masutani C. Kanao R, et al. Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
Cited by
- Readers of PCNA modifications.
Ulrich HD, Takahashi DT. Ulrich HD, et al. Chromosoma. 2013 Aug;122(4):259-74. doi: 10.1007/s00412-013-0410-4. Epub 2013 Apr 12. Chromosoma. 2013. PMID: 23580141 Free PMC article. Review. - Time for remodeling: SNF2-family DNA translocases in replication fork metabolism and human disease.
Joseph SA, Taglialatela A, Leuzzi G, Huang JW, Cuella-Martin R, Ciccia A. Joseph SA, et al. DNA Repair (Amst). 2020 Nov;95:102943. doi: 10.1016/j.dnarep.2020.102943. Epub 2020 Aug 15. DNA Repair (Amst). 2020. PMID: 32971328 Free PMC article. Review. - Reversal of PCNA ubiquitylation by Ubp10 in Saccharomyces cerevisiae.
Gallego-Sánchez A, Andrés S, Conde F, San-Segundo PA, Bueno A. Gallego-Sánchez A, et al. PLoS Genet. 2012;8(7):e1002826. doi: 10.1371/journal.pgen.1002826. Epub 2012 Jul 19. PLoS Genet. 2012. PMID: 22829782 Free PMC article. - Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis.
Haracska L, Unk I, Prakash L, Prakash S. Haracska L, et al. Proc Natl Acad Sci U S A. 2006 Apr 25;103(17):6477-82. doi: 10.1073/pnas.0510924103. Epub 2006 Apr 12. Proc Natl Acad Sci U S A. 2006. PMID: 16611731 Free PMC article. - Phosphorylation Alters the Properties of Pol η: Implications for Translesion Synthesis.
Peddu C, Zhang S, Zhao H, Wong A, Lee EYC, Lee MYWT, Zhang Z. Peddu C, et al. iScience. 2018 Aug 31;6:52-67. doi: 10.1016/j.isci.2018.07.009. Epub 2018 Jul 18. iScience. 2018. PMID: 30240625 Free PMC article.
References
- Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. (2002) Nature 419, 135–141. - PubMed
- Papouli, E., Chen, S., Davies, A. A., Huttner, D., Krejci, L., Sung, P. & Ulrich, H. D. (2005) Mol. Cell 19, 123–133. - PubMed
- Stelter, P. & Ulrich, H. D. (2003) Nature 425, 188–191. - PubMed
- Prakash, S., Johnson, R. E. & Prakash, L. (2005) Annu. Rev. Biochem. 74, 317–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous