Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes - PubMed (original) (raw)
doi: 10.1101/gr.4039406. Epub 2005 Dec 12.
Ai Wakamatsu, Yutaka Suzuki, Toshio Ota, Tetsuo Nishikawa, Riu Yamashita, Jun-ichi Yamamoto, Mitsuo Sekine, Katsuki Tsuritani, Hiroyuki Wakaguri, Shizuko Ishii, Tomoyasu Sugiyama, Kaoru Saito, Yuko Isono, Ryotaro Irie, Norihiro Kushida, Takahiro Yoneyama, Rie Otsuka, Katsuhiro Kanda, Takahide Yokoi, Hiroshi Kondo, Masako Wagatsuma, Katsuji Murakawa, Shinichi Ishida, Tadashi Ishibashi, Asako Takahashi-Fujii, Tomoo Tanase, Keiichi Nagai, Hisashi Kikuchi, Kenta Nakai, Takao Isogai, Sumio Sugano
Affiliations
- PMID: 16344560
- PMCID: PMC1356129
- DOI: 10.1101/gr.4039406
Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes
Kouichi Kimura et al. Genome Res. 2006 Jan.
Abstract
By analyzing 1,780,295 5'-end sequences of human full-length cDNAs derived from 164 kinds of oligo-cap cDNA libraries, we identified 269,774 independent positions of transcriptional start sites (TSSs) for 14,628 human RefSeq genes. These TSSs were clustered into 30,964 clusters that were separated from each other by more than 500 bp and thus are very likely to constitute mutually distinct alternative promoters. To our surprise, at least 7674 (52%) human RefSeq genes were subject to regulation by putative alternative promoters (PAPs). On average, there were 3.1 PAPs per gene, with the composition of one CpG-island-containing promoter per 2.6 CpG-less promoters. In 17% of the PAP-containing loci, tissue-specific use of the PAPs was observed. The richest tissue sources of the tissue-specific PAPs were testis and brain. It was also intriguing that the PAP-containing promoters were enriched in the genes encoding signal transduction-related proteins and were rarer in the genes encoding extracellular proteins, possibly reflecting the varied functional requirement for and the restricted expression of those categories of genes, respectively. The patterns of the first exons were highly diverse as well. On average, there were 7.7 different splicing types of first exons per locus partly produced by the PAPs, suggesting that a wide variety of transcripts can be achieved by this mechanism. Our findings suggest that use of alternate promoters and consequent alternative use of first exons should play a pivotal role in generating the complexity required for the highly elaborated molecular systems in humans.
Figures
Figure 1.
Identification of the putative alternative promoters in human genes. Schematic representation of the mapping of the 5′-ends of the oligo-cap cDNAs, the determination of the TSSs, and clustering of the TSSs to identify the PAPs. The boxes and lines represent exons and introns, respectively. The RefSeq sequences and the oligo-cap cDNAs are in red and blue, respectively. The lowest gray oligo-cap cDNA is excluded from the data set, since its 5′-end is located within an internal exon of the RefSeq. The third-lowest oligo-cap cDNA is accepted because the truncation of the erroneously sliced second-lowest transcript would otherwise need to be hypothesized to explain its presence, and the chance of the combination of such events should be low. The shaded boxes represent the retained introns. Altogether, this case consists of 8 “full-length” oligo-cap cDNAs that are mapped at 6 TSSs, clustered into 3 PAPs.
Figure 2.
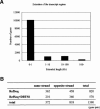
Comparison of the DBTSS data with the previously characterized TSSs and APs. TSSs (A) and APs (B) identified by the DBTSS data were compared with those characterized in previous studies. When a TSS/AP registered in EPD was located within 100 bp of one in DBTSS, they were counted as “overlapping.” The margin of 100 bp was allowed considering fluctuations of the TSSs (Suzuki et al. 2001a). (A) The “overlapping” was counted separately for the TSS data obtained from high-throughput cDNA cloning methods (like ours) and that from conventional methods, such as RACE and nuclease protection assays. Note that as some of the TSSs were identified by multiple methods, the total numbers in the third line are not always the sum of the above two. (B) First column is the total number of EPD genes registered as “alternative promoter-containing genes” and the number of the corresponding promoters; second column is the coverage of the DBTSS against EPD at the gene level; third column is coverage of the DBTSS against EPD at the promoter level (all APs were covered by DBTSS PPRs). (C) The case in which EPD data and DBTSS data were overlapping with each other is exemplified by the case of the human hydroxymethylbilane synthase gene (NM_000190). RefSeq exons are shown in blue (non-coding regions) and yellow (coding regions) boxes and the DBTSS exons are shown in red (PAP group 1) and green (PAP group 2) boxes. The lower panels are magnifications of the upper panel(s). The TSSs are represented by arrows of the corresponding colors. The IDs of corresponding EPD data are shown. Note that there are variations in the first exon patterns even within the same PAP group (alternative donor in PAP1 and retaining intron in PAP2) and the TSSs are fluctuating. For additional examples, see Supplemental Table 3.
Figure 2.
Comparison of the DBTSS data with the previously characterized TSSs and APs. TSSs (A) and APs (B) identified by the DBTSS data were compared with those characterized in previous studies. When a TSS/AP registered in EPD was located within 100 bp of one in DBTSS, they were counted as “overlapping.” The margin of 100 bp was allowed considering fluctuations of the TSSs (Suzuki et al. 2001a). (A) The “overlapping” was counted separately for the TSS data obtained from high-throughput cDNA cloning methods (like ours) and that from conventional methods, such as RACE and nuclease protection assays. Note that as some of the TSSs were identified by multiple methods, the total numbers in the third line are not always the sum of the above two. (B) First column is the total number of EPD genes registered as “alternative promoter-containing genes” and the number of the corresponding promoters; second column is the coverage of the DBTSS against EPD at the gene level; third column is coverage of the DBTSS against EPD at the promoter level (all APs were covered by DBTSS PPRs). (C) The case in which EPD data and DBTSS data were overlapping with each other is exemplified by the case of the human hydroxymethylbilane synthase gene (NM_000190). RefSeq exons are shown in blue (non-coding regions) and yellow (coding regions) boxes and the DBTSS exons are shown in red (PAP group 1) and green (PAP group 2) boxes. The lower panels are magnifications of the upper panel(s). The TSSs are represented by arrows of the corresponding colors. The IDs of corresponding EPD data are shown. Note that there are variations in the first exon patterns even within the same PAP group (alternative donor in PAP1 and retaining intron in PAP2) and the TSSs are fluctuating. For additional examples, see Supplemental Table 3.
Figure 3.
Relationship between the PAPs and the CpG islands and TATA boxes. Frequencies of the CpG island (A) and TATA box (B) containing PPRs. In the right panels, the relationship between the number of PAPs (_x_-axis) and the frequency of the corresponding promoter motif (_y_-axis) is shown.
Figure 4.
Tissue-specific usage of PAPs. (A) The number of PAPs that are used in a tissue-specific manner. For the detailed definition of the tissue specificity, see the Methods section. (B) Examples of tissue-specific PAPs. The _x_-axes represent the genomic positions and the bars represent the number of 5′-ends of the oligo-cap cDNAs mapped at the corresponding genomic positions (TSSs). White bars show the tissue-specific usage of the corresponding PAPs observed in the indicated tissues.
Figure 5.
Patterns of the first exons in the PAP-containing genes. (A) Distributions of the patterns of the first exons are shown. The number of identified exon patterns was counted in total (third column) or between the populations which are separated by >500 bp, thus accounting for the separation of the PAPs. *Either of the first exon variations was a “single exon” transcript. Different criteria were employed for them because these transcripts cannot be regarded as “splicing” variants. (B) Alterations of the amino acids resulting from the exon variations occurring in the population of “inter-APs” (TSS distance >500) or “intra-APs” (TSS distance ≤500) were counted.
Figure 6.
Identification of putative overlapping and anti-sense gene pairs. (A) Length distribution of the RefSeq (LocusLink) regions extended by the additional DBTSS data. (B) Number of putative gene pairs identified using the indicated data set.
Similar articles
- Systematic analysis of alternative promoters correlated with alternative splicing in human genes.
Ma X, Li-Ling J, Huang Q, Chen X, Hou L, Ma F. Ma X, et al. Genomics. 2009 May;93(5):420-5. doi: 10.1016/j.ygeno.2009.01.008. Epub 2009 Feb 11. Genomics. 2009. PMID: 19442634 - Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs.
Amarger V, Nguyen M, Van Laere AS, Braunschweig M, Nezer C, Georges M, Andersson L. Amarger V, et al. Mamm Genome. 2002 Jul;13(7):388-98. doi: 10.1007/s00335-001-3059-x. Mamm Genome. 2002. PMID: 12140686 - Contrasting chromatin organization of CpG islands and exons in the human genome.
Choi JK. Choi JK. Genome Biol. 2010;11(7):R70. doi: 10.1186/gb-2010-11-7-r70. Epub 2010 Jul 5. Genome Biol. 2010. PMID: 20602769 Free PMC article. - Tissue specific glucocorticoid receptor expression, a role for alternative first exon usage?
Turner JD, Schote AB, Macedo JA, Pelascini LP, Muller CP. Turner JD, et al. Biochem Pharmacol. 2006 Nov 30;72(11):1529-37. doi: 10.1016/j.bcp.2006.07.005. Epub 2006 Aug 22. Biochem Pharmacol. 2006. PMID: 16930562 Review.
Cited by
- Retrotransposon-derived promoter of Mammalian Aebp2.
Kim H, Bakshi A, Kim J. Kim H, et al. PLoS One. 2015 Apr 27;10(4):e0126966. doi: 10.1371/journal.pone.0126966. eCollection 2015. PLoS One. 2015. PMID: 25915901 Free PMC article. - Alternate promoter usage generates two subpopulations of the neuronal RhoGEF Kalirin-7.
Miller MB, Yan Y, Wu Y, Hao B, Mains RE, Eipper BA. Miller MB, et al. J Neurochem. 2017 Mar;140(6):889-902. doi: 10.1111/jnc.13749. Epub 2016 Sep 6. J Neurochem. 2017. PMID: 27465683 Free PMC article. - Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination.
Abarrategui I, Krangel MS. Abarrategui I, et al. EMBO J. 2007 Oct 17;26(20):4380-90. doi: 10.1038/sj.emboj.7601866. Epub 2007 Sep 20. EMBO J. 2007. PMID: 17882258 Free PMC article. - Identification and functional analyses of 11,769 full-length human cDNAs focused on alternative splicing.
Wakamatsu A, Kimura K, Yamamoto J, Nishikawa T, Nomura N, Sugano S, Isogai T. Wakamatsu A, et al. DNA Res. 2009 Dec;16(6):371-83. doi: 10.1093/dnares/dsp022. Epub 2009 Oct 30. DNA Res. 2009. PMID: 19880432 Free PMC article. - ALDH1A2 (RALDH2) genetic variation in human congenital heart disease.
Pavan M, Ruiz VF, Silva FA, Sobreira TJ, Cravo RM, Vasconcelos M, Marques LP, Mesquita SM, Krieger JE, Lopes AA, Oliveira PS, Pereira AC, Xavier-Neto J. Pavan M, et al. BMC Med Genet. 2009 Nov 3;10:113. doi: 10.1186/1471-2350-10-113. BMC Med Genet. 2009. PMID: 19886994 Free PMC article.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287 2185-2195. - PubMed
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103 367-370. - PubMed
- Boguski, M.S. 2002. Comparative genomics: The mouse that roared. Nature 420 515-516. - PubMed
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282 2012-2018. - PubMed
Web site references
- http://dbtss.hgc.jp/; DBTSS.
- http://www.genome.gov/10005107/; ENCODE.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous