Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells - PubMed (original) (raw)
Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells
Cheng Huang et al. J Virol. 2006 Jan.
Abstract
Severe acute respiratory syndrome coronavirus (SCoV) accessory protein 3a is a virus structural protein. We demonstrate here that 3a protein was released efficiently in membranous structures from various cell lines expressing 3a protein. A subpopulation of the released 3a protein is associated with detergent-resistant membranes. The presence of the YxxPhi and diacidic motifs, located within the cytoplasmic tail of the 3a protein, was not required for its efficient release. Analysis of supernatant from SCoV-infected cells with sucrose gradient sedimentation and virus capture assay indicated that the 3a protein was released from infected cells in two distinct populations, as a component of SCoV particles, and in membrane structures with a lower buoyant density. These data provide new insights into the biological properties of SCoV 3a protein.
Figures
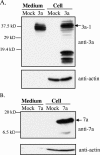
FIG. 1.
Release of SCoV 3a protein from 3a protein-expressing cells. 293T cells were independently transfected with pCAGGS (mock), pCAGGS-3a (3a), or pCAGGS-7a (7a). After 48 h transfection, supernatants from transfected cells were clarified, applied onto a 20% sucrose cushion, and centrifuged at 26,000 rpm for 3 h at 4°C. The pellets were suspended in 1× SDS-PAGE loading buffer (Medium). Cell lysates were prepared with 1× SDS-PAGE loading buffer (Cell). Samples were subjected to Western blotting analysis with anti-3a antibody (A) and anti-7a antibody (B). The membranes were reprobed with anti-actin antibody (anti-actin). 3a-1 indicates 3a protein with a molecular mass of ∼37 kDa.
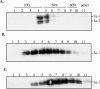
FIG. 2.
Flotation assay of 3a-containing membranous structures. Purified 3a-containing membrane structures from the media of 3a-expressing 293T cells were suspended in NTE buffer (A), in NTE buffer containing 1% Triton X-100 and incubated at 4°C for 30 min (B), or in NTE buffer containing 1% Triton X-100 at room temperature and incubated for 30 min (C). After sucrose was added to adjust the final concentration to 60%, the samples were transferred to the bottom of Beckman SW41 centrifuge tubes and overlaid with 50% sucrose and 10% sucrose. The step gradient was centrifuged to equilibrium at 38,000 rpm for 18 h by using a Beckman SW41 rotor. Fractions were collected from top to bottom. Equal amounts of each fraction were diluted in NTE buffer, separated by SDS-PAGE, and analyzed by Western blotting with anti-3a antibody. 3a-1 and 3a-2 indicate the slow-migrating (37 kDa) and fast-migrating (31 kDa) forms of the 3a protein, respectively.
FIG. 3.
Analysis of the buoyant density of 3a-containing membrane structures. The supernatant of 3a-expressing 293T cells was collected at 48 h after transfection and partially purified by centrifugation on a 20% sucrose cushion. The pellets were suspended in NTE buffer, loaded on a 20 to 60% continuous sucrose gradient, and centrifuged at 26,000 rpm for 18 h using an SW28 rotor. Twelve fractions were taken from the bottom, and the densities of each action were measured (upper panel). Each fraction was then diluted at least threefold with NTE buffer and was then centrifuged on a 20% sucrose cushion. The recovered pellets were suspended in 1× SDS-PAGE loading buffer and subjected to Western blotting analysis with anti-3a antibody (lower panel).
FIG. 4.
Release of 3a protein from SCoV-infected cells. (A) Supernatants from SCoV-infected Caco2 cells were harvested and inactivated by irradiation. The samples were clarified and partially purified by centrifugation on a 20% sucrose cushion. SCoV virions were suspended in NTE buffer, loaded on continuous 20 to 60% sucrose gradient, and centrifuged at 26,000 rpm for 18 h by using a Beckman SW28 rotor. Twelve fractions were taken from the bottom, and each was then diluted at least three times with NTE buffer, followed by centrifugation on 20% sucrose cushion. The recovered virus samples were dissolved in 1× SDS-PAGE loading buffer and detected by Western blotting with anti-3a antibody, anti-SCoV N protein antibody, and anti-SCoV S protein antibody, as indicated. 3a-1 and 3a-2 indicate the slow-migrating form (37 kDa) and fast-migrating form (31 kDa) of 3a protein, respectively. (B) The 3a, N, and S protein levels were measured by densitometry analysis with AlphaEase software (Alpha Innotech). For each protein, the level of protein in fraction 5 was arbitrarily set as 1. The relative amounts of each protein in different fractions are shown.
FIG. 5.
SCoV capture assay. Clarified supernatants from SCoV-infected Caco2 cells were applied to the top of the 20% sucrose cushion, and then the samples were centrifuged at 26,000 rpm for 3 h at 4°C by using a Beckman SW28 rotor. Lane 1 represents the pellet fraction. The suspended pellets were immunoprecipitated by nonspecific monoclonal mouse anti-H-2K antibody (lane 2) or by a mouse monoclonal anti-SCoV S antibody to capture virions (lane 3). Lane 4 represents supernatant that was not precipitated by anti-SCoV S antibody. All of the samples were examined with a rabbit anti-SCoV 3a antibody (A) and a rabbit anti-SCoV M antibody (B) in Western blot analysis.
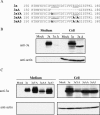
FIG. 6.
Role of YxxΦ and diacidic motifs in the release of 3a protein. (A) Partial sequences of the C-terminal region of 3a, 3aΔ, 3aYA, 3aAA, and 3aA3 proteins. YxxΦ and diacidic motifs are underlined. (B and C) 293T cells were independently transfected with pCAGGS (mock), pCAGGS-3a (3a), pCAGGS-3aΔ (3aΔ), pCAGGS-3aYA (3aYA), pCAGGS-3aAA (3aAA), and pCAGGS-3aA3 (3aA3). After 48 h transfection, supernatants from transfected cells were clarified and purified by centrifugation on a 20% sucrose cushion. The pellets were dissolved in 1× SDS-PAGE loading buffer (Medium). Cell lysates were prepared with 1× SDS-PAGE loading buffer (Cell). Samples were subjected to Western blotting analysis with anti-3a antibody to detect 3a protein. The membranes were reprobed with anti-actin antibody (anti-actin).
Similar articles
- Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein.
Ito N, Mossel EC, Narayanan K, Popov VL, Huang C, Inoue T, Peters CJ, Makino S. Ito N, et al. J Virol. 2005 Mar;79(5):3182-6. doi: 10.1128/JVI.79.5.3182-3186.2005. J Virol. 2005. PMID: 15709039 Free PMC article. - Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis.
Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Verdia-Báguena C, Queralt-Martín M, Kochan G, Perlman S, Aguilella VM, Sola I, Enjuanes L. Castaño-Rodriguez C, et al. mBio. 2018 May 22;9(3):e02325-17. doi: 10.1128/mBio.02325-17. mBio. 2018. PMID: 29789363 Free PMC article. - Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells.
Huang C, Peters CJ, Makino S. Huang C, et al. J Virol. 2007 May;81(10):5423-6. doi: 10.1128/JVI.02307-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344286 Free PMC article. - Expression of the severe acute respiratory syndrome coronavirus 3a protein and the assembly of coronavirus-like particles in the baculovirus expression system.
Khan S, Ng ML, Tan YJ. Khan S, et al. Methods Mol Biol. 2007;379:35-50. doi: 10.1007/978-1-59745-393-6_3. Methods Mol Biol. 2007. PMID: 17502669 Free PMC article. Review. - SARS coronavirus accessory proteins.
Narayanan K, Huang C, Makino S. Narayanan K, et al. Virus Res. 2008 Apr;133(1):113-21. doi: 10.1016/j.virusres.2007.10.009. Epub 2007 Nov 28. Virus Res. 2008. PMID: 18045721 Free PMC article. Review.
Cited by
- Interaction between SARS-CoV PBM and Cellular PDZ Domains Leading to Virus Virulence.
Honrubia JM, Valverde JR, Muñoz-Santos D, Ripoll-Gómez J, de la Blanca N, Izquierdo J, Villarejo-Torres M, Marchena-Pasero A, Rueda-Huélamo M, Nombela I, Ruiz-Yuste M, Zuñiga S, Sola I, Enjuanes L. Honrubia JM, et al. Viruses. 2024 Jul 29;16(8):1214. doi: 10.3390/v16081214. Viruses. 2024. PMID: 39205188 Free PMC article. - Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19.
Zhang J, Ejikemeuwa A, Gerzanich V, Nasr M, Tang Q, Simard JM, Zhao RY. Zhang J, et al. Front Microbiol. 2022 Mar 9;13:854567. doi: 10.3389/fmicb.2022.854567. eCollection 2022. Front Microbiol. 2022. PMID: 35356515 Free PMC article. Review. - D155Y substitution of SARS-CoV-2 ORF3a weakens binding with Caveolin-1.
Gupta S, Mallick D, Banerjee K, Mukherjee S, Sarkar S, Lee ST, Basuchowdhuri P, Jana SS. Gupta S, et al. Comput Struct Biotechnol J. 2022;20:766-778. doi: 10.1016/j.csbj.2022.01.017. Epub 2022 Jan 31. Comput Struct Biotechnol J. 2022. PMID: 35126886 Free PMC article. - Unravelling the Immunomodulatory Effects of Viral Ion Channels, towards the Treatment of Disease.
Gargan S, Stevenson NJ. Gargan S, et al. Viruses. 2021 Oct 27;13(11):2165. doi: 10.3390/v13112165. Viruses. 2021. PMID: 34834972 Free PMC article. Review. - Coronavirus, the King Who Wanted More Than a Crown: From Common to the Highly Pathogenic SARS-CoV-2, Is the Key in the Accessory Genes?
Chazal N. Chazal N. Front Microbiol. 2021 Jul 14;12:682603. doi: 10.3389/fmicb.2021.682603. eCollection 2021. Front Microbiol. 2021. PMID: 34335504 Free PMC article. Review.
References
- Alimonti, J. B., T. B. Ball, and K. R. Fowke. 2003. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J. Gen. Virol. 84:1649-1661. - PubMed
- Bannykh, S. I., N. Nishimura, and W. E. Balch. 1998. Getting into the Golgi. Trends Cell Biol. 8:21-25. - PubMed
- Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. - PubMed
- Briggs, J. A., T. Wilk, and S. D. Fuller. 2003. Do lipid rafts mediate virus assembly and pseudotyping? J. Gen. Virol. 84:757-768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases