Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration - PubMed (original) (raw)
Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration
Wanli W Smith et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Parkinson's disease (PD) is a disorder of movement, cognition, and emotion, and it is characterized pathologically by neuronal degeneration with Lewy bodies, which are cytoplasmic inclusion bodies containing deposits of aggregated proteins. Most PD cases appear to be sporadic, but genetic forms of the disease, caused by mutations in alpha-synuclein, parkin, and other genes, have helped elucidate pathogenesis. Mutations in leucine-rich repeat kinase 2 (LRRK2) cause autosomal-dominant Parkinsonism with clinical features of PD and with pleomorphic pathology including deposits of aggregated protein. To study expression and interactions of LRRK2, we synthesized cDNAs and generated expression constructs coding for human WT and mutant LRRK2 proteins. Expression of full-length LRRK2 in cells in culture suggests that the protein is predominately cytoplasmic, as is endogenous protein by subcellular fractionation. Using coimmunoprecipitation, we find that LRRK2, expressed in cells in culture, interacts with parkin but not with alpha-synuclein, DJ-1, or tau. A small proportion of the cells overexpressing LRRK2 contain protein aggregates, and this proportion is greatly increased by coexpression of parkin. In addition, parkin increases ubiquitination of aggregated protein. Also, mutant LRRK2 causes neuronal degeneration in both SH-SY5Y cells and primary neurons. This cell model may be useful for studies of PD cellular pathogenesis and therapeutics. These findings suggest a gain-of-function mechanism in the pathogenesis of LRRK2-linked PD and suggest that LRRK2 may be involved in a pathogenic pathway with other PD-related proteins such as parkin, which may help illuminate both familial and sporadic PD.
Figures
Fig. 1.
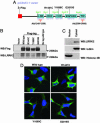
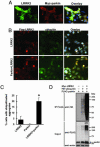
Cytoplasmic expression of LRRK2. (A) Schematic representation of LRRK2 expression construct in pcDNA3.1 vector. The WT construct was designed with or without a FLAG tag. Unique restriction sites were engineered between domains to facilitate cloning. (B) Lysates derived from HEK 293T cells transiently expressing various LRRK2 constructs or empty vector were analyzed by Western blotting using anti-FLAG and anti-LRRK2-(1245-1259) Abs. LRRK2 proteins migrated at ≈280 kDa. Similar results were obtained by using anti-LRRK2-(2396-2408) Ab (data not shown). (C) Subcellular fractionations derived from SH-SY5Y cells were analyzed by Western blot analysis using anti-LRRK2-(1245-1259) Abs to detect the endogenous LRRK2. (D) Representative merged confocal images of transiently transfected HEK 293T cells. Nuclear Hoechst 33342 staining is indicated blue, and green indicates FLAG-FITC Ab staining for LRRK2.
Fig. 2.
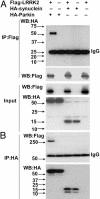
LRRK2 interaction with parkin in HEK 293T cells. (A and B) Lysates prepared from cells transfected with various constructs as indicated were subjected to IP with anti-FLAG (A) or anti-HA (B), followed by anti-HA and anti-FLAG immunoblotting. The experiment was repeated three times with similar results.
Fig. 3.
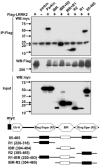
LRRK2 interacts preferentially with the C-terminal R2 ring-finger domain of parkin. Lysates prepared from HEK 293T cells cotransfected with FLAG-LRRK2 and various myc-tagged parkin domain constructs were subjected to IP with anti-FLAG, followed by anti-myc immunoblotting. Putative functional domains of parkin used in the mapping experiments are shown. Ub-H, Ubiquitin homology domain. Cell lysates were subject to Bradford protein assay to ensure equal protein loading. Parkin fragments had variable levels, even when we loaded the same amount of total protein, presumably because these nonphysiological forms are not stable. On a qualitative basis, the RING2 domain of parkin interacts most strongly with LRRK2
Fig. 4.
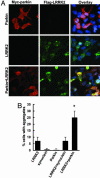
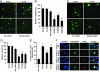
Expression of parkin increases cytoplasmic aggregates containing LRRK2. (A and B) Cells were transfected with WT FLAG-LRRK2 with or without myc-parkin for 72 h and subjected to immunocytochemical assay with anti-myc and anti-FLAG Abs. (A) Representative images of each experimental group. (B) Cells with cytoplasmic aggregates containing FLAG-LRRK2 labeling were counted. Data are shown as means ± SE for three separate experiments performed in duplicate. *, P < 0.05 vs. cells transfected with FLAG-LRRK2 alone.
Fig. 5.
Expression of parkin increases ubiquitinated cytoplasmic aggregates containing LRRK2. (A and B) Cells were transfected with WT FLAG-LRRK2 with or without myc-parkin for 72 h and subjected to immunocytochemical assay with anti-FLAG and anti-myc (A) or anti-FLAG and anti-ubiquitin (B) Abs. (C) Cells with cytoplasmic aggregates containing both ubiquitin and FLAG labeling were counted. Data are shown as means ± SE for three separate experiments performed in duplicate. *, P < 0.05 vs. cells transfected with FLAG-LRRK2 alone. (D) LRRK2 enhances parkin autoubiquitination. Cells cotransfected with FLAG-parkin, HA-ubiquitin, myc-LRRK2, or EGFP plasmid as a control for 48 h and then were subjected to IP with anti-FLAG Ab. Immunoprecipitates were analyzed by Western blotting with anti-HA Ab, and inputs were probed with anti-FLAG or anti-actin Abs. Note that LRRK2 enhances the formation of high-molecular-weight parkin-ubiquitin protein conjugates corresponding to polyubiquitinated forms of parkin. These experiments were replicated three times with similar results.
Fig. 6.
Mutant LRRK2 causes neuronal degeneration. (A) SH-SY5Y cells were cotransfected with pcDNA3.1-GFP along with either vector pcDNA3.1, full-length htt with 23 Q (FL-Htt), WT, or mutant pcDNA3.1-FLAG-LRRK2 at 1:15 ratios for 24 h in 10% FBS OPTI-MEM I media, then incubated with DMEM containing N2 supplement for 24 h. GFP-positive cells with 2-fold continuous extensions were counted by using fluorescence microscopy. Shown are representative photomicrographs for each experimental group. Transfection efficiency of GFP was ≈10%. (B) Quantitation of data in A, representing the cell viability of each experimental group normalized to that of cells cotransfected with empty vector and GFP. The percentage of GFP-positive viable cells in each experimental group relative to those of cells cotransfected with vector and GFP was calculated. *, P < 0.05 vs. cells cotransfected with WT LRRK2 and GFP. (C) Mouse primary cortical neurons were cotransfected with pcDNA3.1-GFP along with various constructs as in A at 1:15 for 48 h by electroporation. GFP-positive viable neurons with 2-fold continuous neurites were counted by using fluorescence microscopy. Shown are representative photomicrographs for each experimental group. (D) Quantitation of data in C, representing the cell viability of each experimental group normalized to that of cells cotransfected with empty vector and GFP as in B. *, P < 0.05 vs. cells cotransfected with WT LRRK2 and GFP. (E and F) SH-SY5Y cells were cotransfected with various constructs for 24 h in 10% FBS OPTI-MEM I media and then incubated with DMEM containing N2 supplement for 24 h, followed by anti-FLAG immunostaining and TUNEL assays. (F) Representative confocal images of transiently transfected SH-SY5Y cells. Blue indicates nuclear Hoechst 33342 staining, green indicates FLAG-FITC Ab staining for LRRK2, and red indicates TUNEL staining. (E) Quantitation of data in F, representing the percentage of TUNEL-positive cells in total LRRK2 transfected cells. *, P < 0.05 vs. cells transfected with WT LRRK2.
Similar articles
- LRRK2 and parkin immunoreactivity in multiple system atrophy inclusions.
Huang Y, Song YJ, Murphy K, Holton JL, Lashley T, Revesz T, Gai WP, Halliday GM. Huang Y, et al. Acta Neuropathol. 2008 Dec;116(6):639-46. doi: 10.1007/s00401-008-0446-3. Epub 2008 Oct 21. Acta Neuropathol. 2008. PMID: 18936941 - Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein.
Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, Chen ZZ, Gallant PE, Tao-Cheng JH, Rudow G, Troncoso JC, Liu Z, Li Z, Cai H. Lin X, et al. Neuron. 2009 Dec 24;64(6):807-27. doi: 10.1016/j.neuron.2009.11.006. Neuron. 2009. PMID: 20064389 Free PMC article. - LRRK2 and neurodegeneration.
Santpere G, Ferrer I. Santpere G, et al. Acta Neuropathol. 2009 Mar;117(3):227-46. doi: 10.1007/s00401-008-0478-8. Epub 2009 Jan 14. Acta Neuropathol. 2009. PMID: 19142648 Review. - Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells.
Plowey ED, Cherra SJ 3rd, Liu YJ, Chu CT. Plowey ED, et al. J Neurochem. 2008 May;105(3):1048-56. doi: 10.1111/j.1471-4159.2008.05217.x. Epub 2008 Jan 7. J Neurochem. 2008. PMID: 18182054 Free PMC article. - [Clinical molecular genetics for PARK8 (LRRK2)].
Tomiyama H, Hatano T, Hattori N. Tomiyama H, et al. Brain Nerve. 2007 Aug;59(8):839-50. Brain Nerve. 2007. PMID: 17713120 Review. Japanese.
Cited by
- Genes Implicated in Familial Parkinson's Disease Provide a Dual Picture of Nigral Dopaminergic Neurodegeneration with Mitochondria Taking Center Stage.
Franco R, Rivas-Santisteban R, Navarro G, Pinna A, Reyes-Resina I. Franco R, et al. Int J Mol Sci. 2021 Apr 28;22(9):4643. doi: 10.3390/ijms22094643. Int J Mol Sci. 2021. PMID: 33924963 Free PMC article. Review. - The impact of genetic research on our understanding of Parkinson's disease.
Martin I, Dawson VL, Dawson TM. Martin I, et al. Prog Brain Res. 2010;183:21-41. doi: 10.1016/S0079-6123(10)83002-X. Prog Brain Res. 2010. PMID: 20696313 Free PMC article. - Development of inducible leucine-rich repeat kinase 2 (LRRK2) cell lines for therapeutics development in Parkinson's disease.
Huang L, Shimoji M, Wang J, Shah S, Kamila S, Biehl ER, Lim S, Chang A, Maguire-Zeiss KA, Su X, Federoff HJ. Huang L, et al. Neurotherapeutics. 2013 Oct;10(4):840-51. doi: 10.1007/s13311-013-0208-3. Neurotherapeutics. 2013. PMID: 23963789 Free PMC article. - Guilt-by-Association - Functional Insights Gained From Studying the LRRK2 Interactome.
Gloeckner CJ, Porras P. Gloeckner CJ, et al. Front Neurosci. 2020 May 20;14:485. doi: 10.3389/fnins.2020.00485. eCollection 2020. Front Neurosci. 2020. PMID: 32508578 Free PMC article. Review. - Fibroblast Biomarkers of Sporadic Parkinson's Disease and LRRK2 Kinase Inhibition.
Smith GA, Jansson J, Rocha EM, Osborn T, Hallett PJ, Isacson O. Smith GA, et al. Mol Neurobiol. 2016 Oct;53(8):5161-77. doi: 10.1007/s12035-015-9435-4. Epub 2015 Sep 23. Mol Neurobiol. 2016. PMID: 26399642 Free PMC article.
References
- Dauer, W. & Przedborski, S. (2003) Neuron 39, 889-909. - PubMed
- Forno, L. S. (1996) J. Neuropathol. Exp. Neurol. 55, 259-272. - PubMed
- Mouradian, M. M. (2002) Neurology 58, 179-185. - PubMed
- Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819-822. - PubMed
- Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991-1995. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials