Kaiso-deficient mice show resistance to intestinal cancer - PubMed (original) (raw)
. 2006 Jan;26(1):199-208.
doi: 10.1128/MCB.26.1.199-208.2006.
Owen Sansom, Jim Selfridge, Isabel M Caballero, Sergey Salozhin, Dana Aithozhina, Leandro Cerchietti, Fan Guo Meng, Leonard H Augenlicht, John M Mariadason, Brian Hendrich, Ari Melnick, Egor Prokhortchouk, Alan Clarke, Adrian Bird
Affiliations
- PMID: 16354691
- PMCID: PMC1317619
- DOI: 10.1128/MCB.26.1.199-208.2006
Kaiso-deficient mice show resistance to intestinal cancer
Anna Prokhortchouk et al. Mol Cell Biol. 2006 Jan.
Abstract
Kaiso is a BTB domain protein that associates with the signaling molecule p120-catenin and binds to the methylated sequence mCGmCG or the nonmethylated sequence CTGCNA to modulate transcription. In Xenopus laevis, xKaiso deficiency leads to embryonic death accompanied by premature gene activation in blastulae and upregulation of the xWnt11 gene. Kaiso has also been proposed to play an essential role in mammalian synapse-specific transcription. We disrupted the Kaiso gene in mice to assess its role in mammalian development. Kaiso-null mice were viable and fertile, with no detectable abnormalities of development or gene expression. However, when crossed with tumor-susceptible Apc(Min/+) mice, Kaiso-null mice showed a delayed onset of intestinal tumorigenesis. Kaiso was found to be upregulated in murine intestinal tumors and is expressed in human colon cancers. Our data suggest that Kaiso plays a role in intestinal cancer and may therefore represent a potential target for therapeutic intervention.
Figures
FIG. 1.
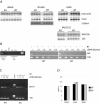
Generation of Kaiso-null mice. (A) Schematic representation of the genomic Kaiso locus, targeting vector, targeted locus, and deleted locus. Boxes represent Kaiso exons that are either translated (black) or untranslated (open). LoxP sites are shown as triangles. The position and direction of primers that were used to validate the targeting are depicted with arrows. (B) Validation of correct targeting by PCR. Primer pairs are indicated on the left. Genomic DNA from a correctly targeted ES clone was used as a template. PCR products were fractionated on 1% agarose gels. DNA fragment sizes are indicated on the right. (C) Northern blot hybridization with Kaiso cDNA. RNA was isolated from brain, kidney, liver, and spleen of wild-type (wt) and _Kaiso_-null (ko) animals. Kaiso mRNA corresponds to ca. 7 kb, and the hybridization signal is indicated by “kaiso.” Prior to the blotting, the gel was stained with ethidium bromide (EtBr) and photographed (bottom panel). (D) Western blot hybridization of liver nuclear extracts from wild-type (wt) and _Kaiso_-null (ko) animals. The Kaiso band (∼100 kDa) and products of Kaiso degradation (from 60 to 45 kDa) are indicated on the left. The bottom panel shows a Western blot of inhibitory κB-alpha protein as an internal control. The protein size markers are on the right. (E) Band-shift assays with nuclear extracts from wt or mutant (ko) liver. The labeled probe was either M+CG11 (methylated) or CG11 (unmethylated). The lower complex in wt M+CG11 lanes is the DNA methylation-specific Kaiso-DNA complex (KGB). This complex is absent in ko lanes. Lanes αKaiso contained anti-Kaiso antibody ZFH6 (33) that specifically supershifts the KGB complex.
FIG.2.
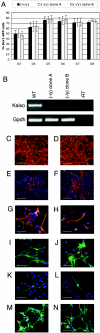
_Kaiso_-null cells show no defects in neural differentiation. (A) Wild-type or Kaiso-deficient ES cells expressing GFP from the Sox1 locus were induced toward neural differentiation for 3, 4, 5, 6, 7, or 8 days (D3, D4, etc.) and fluorescence-activated cell sorted for GFP expression. The experiment was performed in triplicate, with average values (± the standard error of the mean) plotted. (B) Kaiso gene expression was analyzed by RT-PCR in wt and null (clone A and B) sorted Sox1-positive cells. (C to F) Wild-type (C and E) and _Kaiso_-null (D and F) cultures were stained for β-tubulin (C and D) or Gfap (red) and DAPI (4′,6′-diamidino-2-phenylindole; blue) (E and F) after 12 days of monolayer differentiation. (G to N) Wild-type (G, I, K, and M) and _Kaiso_-null (H, J, L, and N) neural stem cells were stained for Nestin (G and H) and RC2 (I and J) or were induced to differentiate and stained for β-tubulin III (K and L) or Gfap (M and N). Cells were counterstained with DAPI (blue). Scale bars: 100 μm (C, D, E, F, M, and N) and 50 μm (G, H, I, J, K, and L).
FIG. 3.
No evidence for abnormal gene expression in _Kaiso_-null animals. (A) Total RNA from four different animals born in two independent families was isolated from wild-type (WT) and _Kaiso_-null (KO) strains. Sources of RNA were brain, liver, spleen, and muscle. The same blot was hybridized with S100A4 and Mta2 probes. Separate blots were prepared with muscle RNA and hybridized with a Rapsyn probe. Normalization of the amount of RNA loaded was performed by reprobing with β-actin (S100A4; Mta2) or S26 ribosomal protein (Rapsyn) probes. (B) In the left panel, chromatin immunoprecipitation was performed with M2 anti-FLAG monoclonal antibodies (M2 lanes) and chromatin was prepared from kidney of wild-type (WT) and _Kaiso_-null (KO) animals. PCR products amplified with _IAP_-specific primers from chromatin immunoprecipitated with or without (“no Ab”) the addition of antibodies are designated. Amplification without DNA (−) and with kidney genomic DNA (+) were used as negative and positive controls, respectively. PCR products were fractionated on 1% agarose gels. In the right panel is shown IAP expression analysis. RNA from liver of wild-type (WT) and _Kaiso_-null (KO) animals was either transcribed (+) or not transcribed (−) by reverse transcriptase (RT). Subsequent PCR amplification of IAP cDNA and control 18S cDNA produced DNA fragments that were resolved on agarose gels. Different dilutions of cDNA were used for PCR amplification as depicted. (C) Semiquantitative RT-PCR with serial dilutions of cDNA from WT or KO heart were amplified by using _Wnt11_-specific primers and compared to 18S rRNA-specific primers used to amplify the same samples. (D) Quantitative “real-time” PCR analysis of Wnt11 mRNA abundance in the hearts, livers, and testes of WT and KO mice. “Delta Ct” expresses the difference in cycle thresholds between Wnt11 and 18S amplification rates.
FIG. 4.
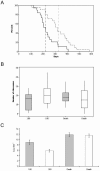
Kaiso deficiency decreases tumor size and increases life span of _Apc_Min/+ mice. (A) Kaplan-Meier survival plot of _Apc_Min/+ Kaiso+/y (solid line) and _Apc_Min/+ _Kaiso_−/y (hashed line) mice. _Apc_Min/+ _Kaiso_−/y mice live significantly longer (median, 317 days; gray vertical line) than both _Apc_Min/+ Kaiso+/y (median, 217; black vertical line, P = 0.006 [log rank]). (B) Tumor number is not altered by Kaiso deficiency. Boxplots show numbers of adenomas per mouse at 180 days and at death. The horizontal boxed line represents the median. Gray boxes, _Apc_Min/+ Kaiso+/y; open boxes, _Apc_Min/+ _Kaiso_−/y. No significant differences were observed between _Apc_Min/+ Kaiso+/y and _Apc_Min/+ _Kaiso_−/y mice at either 180 days (P = 0.10 [Mann-Whitney], n ≥ 20) or at death (P = 0.62 [Mann-Whitney], n ≥ 20). (C) Tumor size, measured by area, is reduced in Kaiso-deficient mice at 180 days (P = 0.001 [Mann-Whitney], n ≥ 114) but not death (P = 0.55 [Mann-Whitney], n ≥ 239). Gray bars, _Apc_Min/+ Kaiso+/y; open bars, _Apc_Min/+ Kaiso −/y. Bars represent the standard error of the mean.
FIG. 5.
Kaiso expression is elevated in murine intestinal tumors. (A) Immunohistochemistry with an anti-Kaiso antibody 6F in colonic tumor and matched normal mucosa from _Muc2_−/− mice at the indicated magnification factors. (B) Western blots performed with anti-Kaiso and antiactin antibodies in two pairs of _Muc2_−/− intestinal tumors compared to matched normal mucosa.
Similar articles
- Epigenetic Regulation of Dlg1, via Kaiso, Alters Mitotic Spindle Polarity and Promotes Intestinal Tumorigenesis.
Young MA, May S, Damo A, Yoon YS, Hur MW, Swat W, Parry L. Young MA, et al. Mol Cancer Res. 2019 Mar;17(3):686-696. doi: 10.1158/1541-7786.MCR-18-0280. Epub 2018 Dec 14. Mol Cancer Res. 2019. PMID: 30552232 - The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development.
Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ, Chekanov N, Li M, Dunican DS, Prokhortchouk E, Pennings S, Meehan RR. Ruzov A, et al. Development. 2009 Mar;136(5):729-38. doi: 10.1242/dev.025569. Epub 2009 Jan 21. Development. 2009. PMID: 19158185 Free PMC article. - The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor.
Daniel JM, Reynolds AB. Daniel JM, et al. Mol Cell Biol. 1999 May;19(5):3614-23. doi: 10.1128/MCB.19.5.3614. Mol Cell Biol. 1999. PMID: 10207085 Free PMC article. - Kaiso Protein in the Regulation of Brain and Behavior.
Kulikova EA, Kulikov AV. Kulikova EA, et al. Curr Protein Pept Sci. 2018;19(7):692-698. doi: 10.2174/1389203718666171030104618. Curr Protein Pept Sci. 2018. PMID: 29086688 Review. - A role for Kaiso-p120ctn complexes in cancer?
van Roy FM, McCrea PD. van Roy FM, et al. Nat Rev Cancer. 2005 Dec;5(12):956-64. doi: 10.1038/nrc1752. Nat Rev Cancer. 2005. PMID: 16294216 Review.
Cited by
- Temporal and epigenetic regulation of neurodevelopmental plasticity.
Allen ND. Allen ND. Philos Trans R Soc Lond B Biol Sci. 2008 Jan 12;363(1489):23-38. doi: 10.1098/rstb.2006.2010. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 17311782 Free PMC article. Review. - POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function.
Lee SU, Maeda T. Lee SU, et al. Immunol Rev. 2012 May;247(1):107-19. doi: 10.1111/j.1600-065X.2012.01116.x. Immunol Rev. 2012. PMID: 22500835 Free PMC article. Review. - Heterozygous loss of Zbtb38 leads to early embryonic lethality via the suppression of Nanog and Sox2 expression.
Nishio M, Matsuura T, Hibi S, Ohta S, Oka C, Sasai N, Ishida Y, Matsuda E. Nishio M, et al. Cell Prolif. 2022 Apr;55(4):e13215. doi: 10.1111/cpr.13215. Epub 2022 Mar 17. Cell Prolif. 2022. PMID: 35297517 Free PMC article. - Cytoplasmic Kaiso is associated with poor prognosis in non-small cell lung cancer.
Dai SD, Wang Y, Miao Y, Zhao Y, Zhang Y, Jiang GY, Zhang PX, Yang ZQ, Wang EH. Dai SD, et al. BMC Cancer. 2009 Jun 9;9:178. doi: 10.1186/1471-2407-9-178. BMC Cancer. 2009. PMID: 19508730 Free PMC article. - Kaiso phosphorylation at threonine 606 leads to its accumulation in the cytoplasm, reducing its transcriptional repression of the tumour suppressor CDH1.
Tian W, Yuan H, Qin S, Liu W, Zhang B, Gu L, Zhou J, Deng D. Tian W, et al. Mol Oncol. 2022 Sep;16(17):3192-3209. doi: 10.1002/1878-0261.13292. Epub 2022 Jul 28. Mol Oncol. 2022. PMID: 35851744 Free PMC article.
References
- Anastasiadis, P. Z., and A. B. Reynolds. 2001. Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 13:604-610. - PubMed
- Antequera, F., J. Boyes, and A. Bird. 1990. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62:503-514. - PubMed
- Chen, W. G., Q. Chang, Y. Lin, A. Meissner, A. E. West, E. C. Griffith, R. Jaenisch, and M. E. Greenberg. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885-889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- G0300058/MRC_/Medical Research Council/United Kingdom
- G0301154/MRC_/Medical Research Council/United Kingdom
- GR067436MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases