Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans - PubMed (original) (raw)
. 2006 Jan;133(2):287-95.
doi: 10.1242/dev.02185. Epub 2005 Dec 14.
Affiliations
- PMID: 16354718
- PMCID: PMC4719054
- DOI: 10.1242/dev.02185
Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans
Florencia Pauli et al. Development. 2006 Jan.
Abstract
We used mRNA tagging to identify genes expressed in the intestine of C. elegans. Animals expressing an epitope-tagged protein that binds the poly-A tail of mRNAs (FLAG::PAB-1) from an intestine-specific promoter (ges-1) were used to immunoprecipitate FLAG::PAB-1/mRNA complexes from the intestine. A total of 1938 intestine-expressed genes (P<0.001) were identified using DNA microarrays. First, we compared the intestine-expressed genes with those expressed in the muscle and germline, and identified 510 genes enriched in all three tissues and 624 intestine-, 230 muscle- and 1135 germ line-enriched genes. Second, we showed that the 1938 intestine-expressed genes were physically clustered on the chromosomes, suggesting that the order of genes in the genome is influenced by the effect of chromatin domains on gene expression. Furthermore, the commonly expressed genes showed more chromosomal clustering than the tissue-enriched genes, suggesting that chromatin domains may influence housekeeping genes more than tissue-specific genes. Third, in order to gain further insight into the regulation of intestinal gene expression, we searched for regulatory motifs. This analysis found that the promoters of the intestine genes were enriched for the GATA transcription factor consensus binding sequence. We experimentally verified these results by showing that the GATA motif is required in cis and that GATA transcription factors are required in trans for expression of these intestinal genes.
Figures
Fig. 1. _ges-1p::_FLAG::PAB-1 is expressed exclusively in the intestine
Immunohistochemistry of strain expressing Pges-1::FLAG::PAB-1 with anti-FLAG antibodies shows intestinal expression in fourth larval stage animals. (A) Bright-field image. (B) Antibody immunoflourescence.
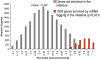
Fig. 2. Genes expressed in the intestine by mRNA tagging
Histogram of the average percentile rank of enrichment after intestinal mRNA tagging. The _x_-axis shows the average percentile rank of enrichment, and the _y_-axis shows the number of genes.
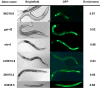
Fig. 3. Intestinal expression of six genes identified by mRNA tagging
The anatomical expression of six genes chosen from the list of 1938 intestinal genes was visualized by transformation with promoter::GFP transcriptional constructs. B0218.8, C25E10.8, ZK970.2, elo-6 and gst-42 show exclusive intestinal expression and D2030.5 shows expression in the intestine as well as other tissues.
Fig. 4. Distribution of intestine-expressed, intestine-, germ line-and muscle-enriched, and commonly-expressed genes on the C. elegans gene expression topomap
The gene expression topomap is an assembly of data from 553 microarray experiments in which genes that are strongly co-expressed are plotted in close proximity to each other on the map (Kim et al., 2001). The gray triangles represent all the genes in the topomap (17,817) and the red triangles represent the genes in each list.
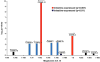
Fig. 5. A chromosomal cluster of intestine-expressed genes
Genes are represented by vertical bars. The distance between genes is indicated on the _x_-axis and the log10(p-value) of enrichment in the intestine by mRNA tagging is indicated on the _y_-axis. Arrows indicate the direction of transcription for each gene.
Fig. 6. The GATA sequence motif is important for intestinal gene expression
(A) Motif logo for the consensus GATA sequence sites identified in 820 out of 1750 intestine-expressed genes by CompareProspector. The _y-_axis represents the information content for each position. (B) Percentage of genes with GATA sites in the intestine and commonly-expressed gene lists. (C) Histogram of the average percentile rank for intestine enrichment of intestine-expressed genes with 0, 1, 2 or 3+ GATA sequence sites.
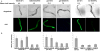
Fig. 7. The GATA sequence is necessary for intestinal gene expression
The GATA sequence in the promoters of elo-6, gst-42 and D2030.5 was mutated and used to generate transgenic GFP reporter strains. (+) indicates the wild-type promoter for each gene and (GATA) indicates that one copy of the GATA motif in the promoter was mutated. Mean pixel intensity was calculated by averaging the image pixel intensity of 20 young adult hermaphrodite animals. (A) Bright-field and GFP images of wild-type and mutant strains. (B) Quantification of GFP expression indicates that the variation between lines is less than the variation between wild-type and mutant strains (P<0.001), indicating that the difference between wild-type and GATA mutant expression levels is not due to variability in the transformation procedure. There is little difference in expression between different lines formed from the same DNA construct, not only in the five lines shown here, but also for three other lines used in a different study (Y.L., unpublished).
Similar articles
- The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine.
McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. McGhee JD, et al. Dev Biol. 2007 Feb 15;302(2):627-45. doi: 10.1016/j.ydbio.2006.10.024. Epub 2006 Oct 21. Dev Biol. 2007. PMID: 17113066 - Multiple cis elements and GATA factors regulate a cuticle collagen gene in Caenorhabditis elegans.
Yin J, Madaan U, Park A, Aftab N, Savage-Dunn C. Yin J, et al. Genesis. 2015 Mar-Apr;53(3-4):278-84. doi: 10.1002/dvg.22847. Epub 2015 Mar 11. Genesis. 2015. PMID: 25711168 Free PMC article. - The function and regulation of the GATA factor ELT-2 in the C. elegans endoderm.
Wiesenfahrt T, Berg JY, Osborne Nishimura E, Robinson AG, Goszczynski B, Lieb JD, McGhee JD. Wiesenfahrt T, et al. Development. 2016 Feb 1;143(3):483-91. doi: 10.1242/dev.130914. Epub 2015 Dec 23. Development. 2016. PMID: 26700680 Free PMC article. - The Caenorhabditis elegans intestine.
McGhee JD. McGhee JD. Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review. - Development of the C. elegans digestive tract.
Kormish JD, Gaudet J, McGhee JD. Kormish JD, et al. Curr Opin Genet Dev. 2010 Aug;20(4):346-54. doi: 10.1016/j.gde.2010.04.012. Epub 2010 Jun 1. Curr Opin Genet Dev. 2010. PMID: 20570129 Review.
Cited by
- Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3'-end-seq.
Haenni S, Ji Z, Hoque M, Rust N, Sharpe H, Eberhard R, Browne C, Hengartner MO, Mellor J, Tian B, Furger A. Haenni S, et al. Nucleic Acids Res. 2012 Jul;40(13):6304-18. doi: 10.1093/nar/gks282. Epub 2012 Mar 29. Nucleic Acids Res. 2012. PMID: 22467213 Free PMC article. - Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network.
Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Owraghi M, et al. Dev Biol. 2010 Apr 15;340(2):209-21. doi: 10.1016/j.ydbio.2009.09.042. Epub 2009 Oct 7. Dev Biol. 2010. PMID: 19818340 Free PMC article. - Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25.
Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, Faergeman NJ, Van Gilst MR, Ingraham HA, Ashrafi K. Mullaney BC, et al. Cell Metab. 2010 Oct 6;12(4):398-410. doi: 10.1016/j.cmet.2010.08.013. Cell Metab. 2010. PMID: 20889131 Free PMC article. - Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection.
Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Wong D, et al. Genome Biol. 2007;8(9):R194. doi: 10.1186/gb-2007-8-9-r194. Genome Biol. 2007. PMID: 17875205 Free PMC article. - Gut neuroendocrine signaling regulates synaptic assembly in C. elegans.
Shi Y, Qin L, Wu M, Zheng J, Xie T, Shao Z. Shi Y, et al. EMBO Rep. 2022 Aug 3;23(8):e53267. doi: 10.15252/embr.202153267. Epub 2022 Jun 24. EMBO Rep. 2022. PMID: 35748387 Free PMC article.
References
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. - PubMed
- Christensen M, Estevez A, Yin X, Fox R, Morrison R, McDonnell M, Gleason C, Miller DM, 3rd, Strange K. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron. 2002;33:503–514. - PubMed
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. - PubMed
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG025941/AG/NIA NIH HHS/United States
- R01 GM043977/GM/NIGMS NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- T32 HG00044/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases