Revealing posttranscriptional regulatory elements through network-level conservation - PubMed (original) (raw)
Revealing posttranscriptional regulatory elements through network-level conservation
Chang S Chan et al. PLoS Comput Biol. 2005 Dec.
Abstract
We used network-level conservation between pairs of fly (Drosophila melanogaster/D. pseudoobscura) and worm (Caenorhabditis elegans/C. briggsae) genomes to detect highly conserved mRNA motifs in 3' untranslated regions. Many of these elements are complementary to the 5' extremity of known microRNAs (miRNAs), and likely correspond to their target sites. We also identify known targets of RNA-binding proteins, and many novel sites not yet known to be functional. Coherent sets of genes with similar function often bear the same conserved elements, providing new insights into their cellular functions. We also show that target sites for distinct miRNAs are often simultaneously conserved, suggesting combinatorial regulation by multiple miRNAs. A genome-wide search for conserved stem-loops, containing complementary sequences to the novel sites, revealed many new candidate miRNAs that likely target them. We also provide evidence that posttranscriptional networks have undergone extensive rewiring across distant phyla, despite strong conservation of regulatory elements themselves.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
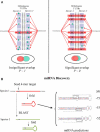
Figure 1. Schematic Representation of the Approach
(A) In the first stage of our approach, we scored exhaustive lists of _k_-mers for network level conservation. Schematic examples for a nonconserved _k_-mer (AAAAAAA) and a highly conserved one (UGUGAUA) are given in the left and right graphics, respectively. (B) In the miRNA discovery stage, seed _k_-mers are used to search the genome for conserved and stable stem-loops.
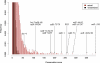
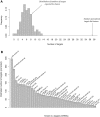
Figure 2. Distribution of Conservation Scores for the C. elegans/C. briggsae Analysis on 3′UTR Sequences
Distributions of actual (red) and randomized (black) sequences are shown. Scores corresponding to some of the known miRNA target sites and RNA-binding protein sites in worms are indicated by arrows. The top portion of both distributions are not shown, for the purpose of presentation.
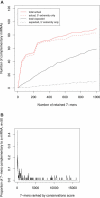
Figure 3. High-Scoring _k_-Mers Are Complementary to the 5′ Ends of Many miRNAs
(A) Number of complementary worm miRNAs as a function of initial number of retained 7-mers. Solid lines correspond to complementarity anywhere within the miRNAs. Dashed lines correspond to complementarity to the 5′ extremity of miRNAs only. Complementarity to the 5′ extremity of a miRNA is defined as starting within 1 nt of the actual miRNA 5′ extremity. (B) Proportion of 7-mers complementary to the 5′ extremity of at least one miRNA, as a function of the conservation rank (using a sliding window [w] of size 50).
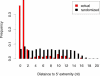
Figure 4. Distribution of Distances from the First Nucleotide of the _k_-Mer to the 5′ Extremity of the miRNA
Distances are given for all pairs of high-scoring _k_-mers/complementary miRNAs. The distribution clearly shows that complementarity between high-scoring worm _k_-mers and miRNAs occurs primarily at the 5′ extremity of the miRNAs.
Figure 5. Number of miRNA Targets
(A) Example showing that the number of predicted targets for D. melanogaster bantam is much larger than expected by chance. The number of predicted targets is the number of genes whose 3′UTR contains at least one conserved _k_-mer complementary to the 5′ extremity of the corresponding miRNA. The distribution of numbers of targets expected by chance was obtained by running the same analysis using 100 pairs of randomized genomes with the same level of divergence as the original ones (see Materials and Methods for details). (B) Estimated numbers of targets for C. elegans miRNAs (only for miRNAs that are complementary to at least one of our high-scoring _k_-mers). Each number corresponds to the number of predicted targets (as defined above) minus the average number of targets expected by chance over the 100 randomizations. The error bars correspond to two standard deviations.
Similar articles
- Identifications of conserved 7-mers in 3'-UTRs and microRNAs in Drosophila.
Gu J, Fu H, Zhang X, Li Y. Gu J, et al. BMC Bioinformatics. 2007 Nov 8;8:432. doi: 10.1186/1471-2105-8-432. BMC Bioinformatics. 2007. PMID: 17996040 Free PMC article. - A conserved sequence motif in 3' untranslated regions of ribosomal protein mRNAs in nematodes.
Hajarnavis A, Durbin R. Hajarnavis A, et al. RNA. 2006 Oct;12(10):1786-9. doi: 10.1261/rna.51306. Epub 2006 Aug 17. RNA. 2006. PMID: 16917125 Free PMC article. - MicroRNA targets in Drosophila.
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Enright AJ, et al. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. Epub 2003 Dec 12. Genome Biol. 2003. PMID: 14709173 Free PMC article. - Translating the Untranslated Region.
Schwerk J, Savan R. Schwerk J, et al. J Immunol. 2015 Oct 1;195(7):2963-71. doi: 10.4049/jimmunol.1500756. J Immunol. 2015. PMID: 26386038 Free PMC article. Review. - Small but influential: the role of microRNAs on gene regulatory network and 3'UTR evolution.
Zhang R, Su B. Zhang R, et al. J Genet Genomics. 2009 Jan;36(1):1-6. doi: 10.1016/S1673-8527(09)60001-1. J Genet Genomics. 2009. PMID: 19161940 Review.
Cited by
- Identification of transcription factor and microRNA binding sites in responsible to fetal alcohol syndrome.
Wang G, Wang X, Wang Y, Yang JY, Li L, Nephew KP, Edenberg HJ, Zhou FC, Liu Y. Wang G, et al. BMC Genomics. 2008;9 Suppl 1(Suppl 1):S19. doi: 10.1186/1471-2164-9-S1-S19. BMC Genomics. 2008. PMID: 18366608 Free PMC article. - The Role of microRNAs in the Repeated Parallel Diversification of Lineages of Midas Cichlid Fish from Nicaragua.
Franchini P, Xiong P, Fruciano C, Meyer A. Franchini P, et al. Genome Biol Evol. 2016 Jun 3;8(5):1543-55. doi: 10.1093/gbe/evw097. Genome Biol Evol. 2016. PMID: 27189980 Free PMC article. - The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression.
Hon LS, Zhang Z. Hon LS, et al. Genome Biol. 2007;8(8):R166. doi: 10.1186/gb-2007-8-8-r166. Genome Biol. 2007. PMID: 17697356 Free PMC article. - Efficient use of accessibility in microRNA target prediction.
Marín RM, Vanícek J. Marín RM, et al. Nucleic Acids Res. 2011 Jan;39(1):19-29. doi: 10.1093/nar/gkq768. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805242 Free PMC article. - Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets.
Morris AR, Mukherjee N, Keene JD. Morris AR, et al. Mol Cell Biol. 2008 Jun;28(12):4093-103. doi: 10.1128/MCB.00155-08. Epub 2008 Apr 14. Mol Cell Biol. 2008. PMID: 18411299 Free PMC article.
References
- Lee R, Feinbaum R, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Hutvagner G, Zamore P. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. - PubMed
- Rhoades M, Reinhart B, Lim L, Burge C, Bartel B, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. - PubMed
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases