Virus membrane-fusion proteins: more than one way to make a hairpin - PubMed (original) (raw)
Review
Virus membrane-fusion proteins: more than one way to make a hairpin
Margaret Kielian et al. Nat Rev Microbiol. 2006 Jan.
Abstract
Structure-function studies have defined two classes of viral membrane-fusion proteins that have radically different architectures but adopt a similar overall 'hairpin' conformation to induce fusion of the viral and cellular membranes and therefore initiate infection. In both classes, the hairpin conformation is achieved after a conformational change is triggered by interaction with the target cell. This review will focus in particular on the properties of the more recently described class II proteins.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
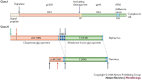
Figure 1. Linear diagrams of the class I and class II fusion proteins.
The nomenclature and organization of the class I proteins of influenza virus and HIV and the class II proteins of flaviviruses and alphaviruses are indicated. For class I, labels specify the positions of the fusion peptides (FP), processing sites (arrows) and transmembrane regions (TM). For class II, signalase cleavage sites are indicated by black arrows, furin-processing sites by red arrows, the capsid protease cleavage site by a green arrow and TM regions are light blue. The intervening 6K segment is unique to alphaviruses. Residue numbers are in parentheses.
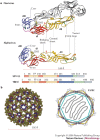
Figure 2. Class II virus membrane-fusion proteins.
a | Structure and sequence diagram of class II virus membrane-fusion proteins. The tick-borne encephalitis (TBE) flavivirus glycoprotein E and the Semliki Forest alphavirus (SFV) E1 protein are shown, with domains I, II and III represented in red, yellow and blue, respectively (dI, dII and dIII). Common elements and important loops in domain II are indicated. Red arrowheads point to the hinge region between domains I and II. The fusion loop is shown in orange, disulphide bonds as green tubes, and carbohydrates (CHO) in pale yellow. The flavivirus E protein dimer is indicated with one subunit in white. b | The icosahedral scaffold formed by E1 at the SFV surface. The left panel shows the lattice resulting from fitting the crystallographic model of E1 on the cryo-electron microscopy reconstruction of the SFV particle. The lattice is formed essentially by E1 interactions around the five-fold, quasi six-fold and quasi two-fold axes of the T = 4 icosahedral particle. All three-fold and quasi three-fold contacts are made exclusively by E2, within the trimeric spike depicted in Fig. 3b. The right panel shows a cartoon of the organization of the SFV particle, with E1 coloured as in Fig. 2a, E2 depicted in pale blue, and the transmembrane (TM) regions drawn as bars crossing the grey lipid bilayer. The internal icosahedral nucleocapsid is indicated by a blue polyhedron with the genomic RNA ('R') inside. Figure prepared using the RIBBONS program.
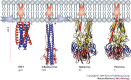
Figure 3. Pre-fusion complexes of class I and II viruses in the conformation present at the surface of infectious particles.
a | shows the influenza virus haemagglutinin (HA1/HA2)3 complex with the HA1 receptor-binding subunits shown in white and the HA2 membrane-fusion subunits coloured in blue and red. The fusion peptide (yellow, residues 1 to 22) is buried at the trimer centre (arrow). b | shows the alphavirus Semliki Forest virus pre-fusion complex (E1/E2)3. The E1 membrane-fusion subunits are drawn as tubes coloured by domains as in Fig. 2. The receptor-binding subunit, E2, is displayed as a sphere-filling model coloured grey, based on subtracting the E1 density from the cryo-electron microscopy reconstruction of the virus. The orange E1 fusion peptide (arrows) is buried at the E1–E2 interface. c | shows the pre-fusion complex (E)2 of the flavivirus tick-borne encephalitis virus. The fusion peptide is buried at the E homodimer interface (arrows). All three panels are drawn at the same scale. The viral membrane would be tangential to a horizontal plane below the drawings. Figure prepared using the RIBBONS program.
Figure 4. Post-fusion conformations of class I and class II fusion proteins.
The class I fusion proteins of HIV-1 and influenza virus (left), and the class II fusion proteins of Semliki Forest virus and tick-borne encephalitis virus (right) are shown. To demonstrate the suggested membrane interactions of the proteins, the final fused membrane is diagrammed in cartoon form. The class I proteins are drawn such that the N-terminal half of the hairpin is blue and the C-terminal part is red, with the missing fusion peptides and transmembrane domains indicated for one of the three trimer subunits as blue and red stars, respectively. For HA2, the residues are coloured as in the neutral pH form displayed in Fig. 3, and a membrane-proximal extended C-terminal 'leash' interacts with the central coiled coil of HA. In HIV glycoprotein 41 (gp41), the post-fusion structure is a six-helix bundle. The inhibitor T20 interacts at the site where the red helix is, effectively inhibiting the fusogenic conformational change. For the class II proteins, an elongated red star indicates the predicted location of the TM segment, with a red arrow pointing to the C terminus of one of the subunits in the trimer of the crystallized class II ectodomains, roughly showing the path of the missing 'stem' regions to complete the protein hairpin. Figure prepared using the RIBBONS program.
Figure 5. Lateral interactions between adjacent Semliki Forest virus E1 trimers.
The left panel shows a negatively stained sample of E1 ectodomain homotrimers (E1*HT) reconstituted at a low lipid-to-protein ratio to produce a planar hexagonal lattice. On liposomes, the E1*HT is proposed to display a fullerene-like architecture, in which insertion of rings of five instead of six trimers allows the required curvature to form a closed sphere (see text). Images of the planar lattice such as that shown on the left were used to generate a three-dimensional reconstruction into which the atomic model of the E1*HT was fitted (right panel). Direct lateral interactions between the trimer heads are observed from the fitting, as shown in this top view. Interactions between the fusion loops of adjacent trimers could also occur through adjustment of the hinge region (indicated in Fig. 2a). Figure prepared using the RIBBONS program.
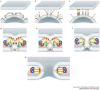
Figure 6. Model for class II membrane fusion, illustrated for Semliki Forest virus.
a | Native virions, showing E1 with domains coloured as in Fig. 2, and E2 in light grey interacting with E1 and covering the fusion loop. Only the target-cell-proximal side of the virion represented in Fig. 2b, right panel, is shown. b | Trigger 1: low pH triggers E1–E2 dissociation and exposure of the fusion loop. c | Trigger 2: low-pH- and cholesterol-dependent insertion of the fusion loop, leading to the alignment of E1 subunits parallel to each other, favouring trimerization. d | Fold-back process: domain III and the stem region move towards the fusion loop. Cooperative interactions between trimers through their fusion loops distort the target membrane. e | Folding of the domain III and stem segments against the body of the trimer pulls transmembrane (TM) segments against each other, distorting the viral membrane. f | Opposing dome-like deformations in the two membranes lead to mixing of the outer leaflets (hemifusion). g | To reach the final stable conformation, the TM segments have to be closely juxtaposed to the fusion loop. This is only possible by opening an initial fusion pore.
Similar articles
- The structural biology of type I viral membrane fusion.
Colman PM, Lawrence MC. Colman PM, et al. Nat Rev Mol Cell Biol. 2003 Apr;4(4):309-19. doi: 10.1038/nrm1076. Nat Rev Mol Cell Biol. 2003. PMID: 12671653 Review. - The Role of histidine residues in low-pH-mediated viral membrane fusion.
Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. Kampmann T, et al. Structure. 2006 Oct;14(10):1481-7. doi: 10.1016/j.str.2006.07.011. Structure. 2006. PMID: 17027497 Review. - Receptor-activated binding of viral fusion proteins to target membranes.
Earp LJ, Hernandez LD, Delos SE, White JM. Earp LJ, et al. Methods Enzymol. 2003;372:428-40. doi: 10.1016/S0076-6879(03)72026-6. Methods Enzymol. 2003. PMID: 14610829 Free PMC article. - Virus membrane fusion.
Weissenhorn W, Hinz A, Gaudin Y. Weissenhorn W, et al. FEBS Lett. 2007 May 22;581(11):2150-5. doi: 10.1016/j.febslet.2007.01.093. Epub 2007 Feb 16. FEBS Lett. 2007. PMID: 17320081 Free PMC article. Review. - Entry mechanisms of enveloped viruses. Implications for fusion of intracellular membranes.
Hoekstra D, Kok JW. Hoekstra D, et al. Biosci Rep. 1989 Jun;9(3):273-305. doi: 10.1007/BF01114682. Biosci Rep. 1989. PMID: 2673423 Review.
Cited by
- Mutagenesis of the DI/DIII linker in dengue virus envelope protein impairs viral particle assembly.
de Wispelaere M, Yang PL. de Wispelaere M, et al. J Virol. 2012 Jul;86(13):7072-83. doi: 10.1128/JVI.00224-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532681 Free PMC article. - Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein.
Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. Krzyzaniak MA, et al. PLoS Pathog. 2013;9(4):e1003309. doi: 10.1371/journal.ppat.1003309. Epub 2013 Apr 11. PLoS Pathog. 2013. PMID: 23593008 Free PMC article. - Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B.
Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Hannah BP, et al. J Virol. 2007 May;81(9):4858-65. doi: 10.1128/JVI.02755-06. Epub 2007 Feb 21. J Virol. 2007. PMID: 17314168 Free PMC article. - Glucosylceramide is essential for Heartland and Dabie bandavirus glycoprotein-induced membrane fusion.
Xia T, Wu X, Hong E, Jung K, Lai CJ, Kwak MJ, Seo H, Kim S, Jiang Z, Cha I, Jung JU. Xia T, et al. PLoS Pathog. 2023 Mar 15;19(3):e1011232. doi: 10.1371/journal.ppat.1011232. eCollection 2023 Mar. PLoS Pathog. 2023. PMID: 36920967 Free PMC article. - Functional Study of the C-Terminal Part of the Hepatitis C Virus E1 Ectodomain.
Moustafa RI, Haddad JG, Linna L, Hanoulle X, Descamps V, Mesalam AA, Baumert TF, Duverlie G, Meuleman P, Dubuisson J, Lavie M. Moustafa RI, et al. J Virol. 2018 Sep 26;92(20):e00939-18. doi: 10.1128/JVI.00939-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068644 Free PMC article.
References
- Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. - PubMed
- Sollner TH. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. - PubMed
- Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. - PubMed
- Sieczkarski SB, Whittaker GR. Viral entry. Curr. Topics Microbiol. Immunol. 2005;285:1–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources