The integral membrane protein Pom34p functionally links nucleoporin subcomplexes - PubMed (original) (raw)
The integral membrane protein Pom34p functionally links nucleoporin subcomplexes
Mi Miao et al. Genetics. 2006 Mar.
Abstract
Here we have examined the function of Pom34p, a novel membrane protein in Saccharomyces cerevisiae, localized to nuclear pore complexes (NPCs). Membrane topology analysis revealed that Pom34p is a double-pass transmembrane protein with both the amino (N) and carboxy (C) termini positioned on the cytosolic/pore face. The network of genetic interactions between POM34 and genes encoding other nucleoporins was established and showed specific links between Pom34p function and Nup170p, Nup188p, Nup59p, Gle2p, Nup159p, and Nup82p. The transmembrane domains of Pom34p in addition to either the N- or C-terminal region were necessary for its function in different double mutants. We further characterized the pom34deltaN nup188delta mutant and found it to be perturbed in both NPC structure and function. Mislocalization of a subset of nucleoporins harboring phenylalanine-glycine repeats was observed, and nuclear import capacity for the Kap104p and Kap121p pathways was inhibited. In contrast, the pom34delta pom152delta double mutant was viable at all temperatures and showed no such defects. Interestingly, POM152 overexpression suppressed the synthetic lethality of pom34delta nup170delta and pom34delta nup59delta mutants. We speculate that multiple integral membrane proteins, either within the nuclear pore domain or in the nuclear envelope, execute coordinated roles in NPC structure and function.
Figures
Figure 1.
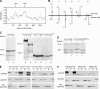
Pom34p is a double-pass integral membrane protein with the N- and the C-terminal regions exposed to the cytosol/pore. (A) Kyte–Doolittle hydropathy analysis (with a 20-amino-acid window size) (K
yte
and D
oolittle
- of Pom34p revealed two spans (arrows) extending from amino acid residues 64 to 84 (TM1) and 132 to 153 (TM2) with significant hydropathic character and length to be transmembrane segments. (B) Six possible topological models for Pom34p relative to the pore membrane are shown. N, amino; C, carboxy. (C) Cell extracts from strains expressing Snl1p-Suc2p (pSW939) (lanes 1 and 2), Pom152p-Suc2pmyc (pSW3192) (lanes 3 and 4), Suc2pmyc-Pom34p (pSW3189) (lanes 5 and 6), or Pom34p-Suc2pmyc (pSW3190) (lanes 7 and 8) were either mock digested (−) or treated with Endo H (+) and analyzed by immunoblotting using rabbit anti-Snl1p (lanes 1 and 2) or mouse monoclonal 9E10 (anti-c-Myc) (lanes 3–8). Molecular mass markers are indicated in kilodaltons. (D) Four potential N-linked glycosylation sites were generated between TM1 and TM2 of Pom34p by site-directed mutagenesis. Cell extracts from SWY2565 expressing either wild-type myc-Pom34p (lanes 1 and 2) or glycosylation site myc-Pom34p-(NXT/S)4 (lanes 3 and 4) were treated with Endo H as in C. Immunoblotting was conducted with the mouse monoclonal 9E10 (anti-c-Myc) antibody. (E) Spheroplast lysates from the _pom34_Δ strain expressing Suc2pmyc-Pom34p (pSW3189), Pom34p-Suc2pmyc (pSW3190), wild-type myc-Pom34p (pSW3193), or myc-Pom34p-(NXT/S)4 (pSW3194) were alkali extracted and separated into supernatant (S indicates S1 and S2) and pellet (P indicates P2) fractions by centrifugation. Fractions were analyzed by immunoblotting with the mouse monoclonal 9E10 (anti-Myc), rabbit anti-Snl1p or with rabbit anti-Nup145C antibodies. (F) Spheroplast lysates from _pom34_Δ cells expressing Pom34p-PrA, TM1Δ-PrA (lacking the TM1), or TM2Δ-PrA (lacking the TM2) were alkali extracted and separated into S and P fractions by centrifugation as in E. Fractions were analyzed by immunoblotting with rabbit anti-mouse (anti-protein A), rabbit anti-Snl1p, or rabbit anti-Nup145C antibodies.
Figure 2.
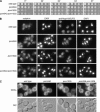
The _pom34_Δ _pom152_Δ double mutant has no significant growth or morphological defects. (A) Fivefold serial dilutions of equal cell numbers from wild type (SWY518), _pom34_Δ (SWY2565), _pom152_Δ (PMY17), or _pom34_Δ _pom152_Δ (SWY3093) strains were spotted on YPD medium. Growth was compared after incubation at the designated temperature. (B) Indirect immuofluorescence microscopy was performed as described in
materials and methods
with either mouse monoclonal mAb414 or rabbit anti-Nup116GLFG antibodies. Nuclei were detected by DAPI staining. (C) Direct fluorescence microscopy of live cells stained with DiOC6 to visualize membranes. Corresponding DIC pictures are shown.
Figure 3.
Multicopy suppression analysis of respective double mutants with POM34 and POM152 expression. (A) _pom34_Δ nup double-mutant strains harboring a POM34/URA3/CEN plasmid (SWY3139, SWY3132, and SWY3125) were transformed with POM152/LEU2/CEN (low copy), _POM152/LEU2/2_μ (high copy), POM34/LEU2/CEN, _POM34/LEU2/2_μ, or empty vectors. The resulting strains were patched on SM-leu and grown at 30° for 2 days before replica plating to 5-FOA medium. The cells on 5-FOA were incubated at 30° for 2 or 7 days (d). (B) _pom152_Δ nup double-mutant strains harboring a POM152/URA3/CEN plasmid (SWY3488, psl21, SWY3219) were transformed with the same LEU2 plasmids, patched, and grown as in A.
Figure 4.
Functional analysis of Pom34p domains. (A) Schematics of the Pom34p structural regions and related deletion/truncation polypeptides are shown. (B) Synthetic lethal double mutants harboring a POM34/URA3/CEN plasmid (low copy) were transformed with the wild-type _POM34/LEU2/2_μ plasmid, the respective mutant pom34/_LEU2/2_μ plasmids (high copy), and empty vector. Transformants were patched on SM-leu (top) at 30° (lanes 1–3) or 23° (lanes 4–6) for 2 days and then replica plated onto 5-FOA media (bottom) and incubated for the indicated days at the same temperature.
Figure 5.
FG Nups mislocalize in the _pom34_Δ_N nup188_Δ mutant strain. Cells in early log phase were processed for indirect immunofluorescence microscopy with mouse monoclonal mAb414, rabbit anti-Nup116GLFG, or mouse monoclonal mAb118C3 (anti-Pom152p) antibodies. Nuclei were detected by DAPI staining.
Figure 6.
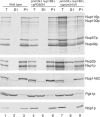
Subcellular fractionation of the _pom34_Δ_N nup188_Δ mutant shows increased levels of FG Nups in the cytoplasm. Spheroplast lysates from wild type (SWY518), _pom34_Δ nup188_Δ + p_POM34 (SWY3489), and _pom34_Δ nup188_Δ + p_pom34_Δ_N (SWY3490) were prepared and separated into soluble (S1) and insoluble pellet (P1) fractions at a low speed centrifugation. Fractions were analyzed by immunoblotting with rabbit anti-Nup116GLFG, rabbit anti-Nsp1p, rabbit anti-Nup145C, mouse monoclonal mAb22C5 (anti-cytoplasmic protein Pgk1p), and mouse monoclonal mAb D77 (anti-nucleolar protein Nop1p) antibodies. The anti-Nup116GLFG antibody recognizes Nup116p, Nup100p, Nup57p, and Nup49p. The anti-Nsp1p antibody recognizes Nup2p in addition to Nsp1p.
Figure 7.
Analysis of nucleocytoplasmic transport in the _pom34_Δ_N nup188_Δ mutant reveals defects in Kap121p and Kap104p import. Cells growing at 30° in early log phase were visualized by direct fluorescence microscopy for steady-state localization of GFP fluorescence. The four GFP reporters analyzed include NLS-GFP (for Kap95p import), NLS-NES-GFP2 (for Xpo1p export), Pho4-NLS-GFP3 (for Kap121p), and Nab2-NLS-GFP (for Kap104p). The corresponding DIC images are shown.
Figure 8.
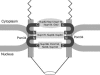
Model for Pom34p in NPC structural organization. The schematic diagram is based on information from published biochemical studies and the results in this article. Connections are shown between the N-terminal region of Pom34p and the Nup170 subcomplex and between the C-terminal Pom34p region and both the Nup188 subcomplex and the Nup82 subcomplex. Pom34p, the Nup170 subcomplex, and the Nup188 subcomplex are all found symmetrically localized on both faces of the NPC (R
out
et al. 2000). The predicted relative stoichiometries of Pom34p and the Nup170 subcomplex are 32 copies/NPC vs. 16 copies/NPC for the Nup188 subcomplex (R
out
et al. 2000). Nic96p and Nsp1p have been isolated in multiple biochemical subcomplexes and are also abundant NPC components at 32 copies/NPC [Nic96p in the Nup188 subcomplex and the Nic96 subcomplex (not shown); Nsp1p in the Nup82 subcomplex and the Nic96 subcomplex (not shown) (reviewed in S
untharalingam
and W
ente
2003)]. The Nup82 complex is exclusively cytoplasmic and predicted to be present in only 8 or 16 copies/NPC. Three peripheral FG Nups are shown assembled in the Nup82 subcomplex (Nsp1p, Nup116p, and Nup159p). Other FG Nups likely assemble by association with these core subcomplexes and the Nup84 subcomplex (not shown).
Similar articles
- The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly.
Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K, Wozniak RW. Makio T, et al. J Cell Biol. 2009 May 4;185(3):459-73. doi: 10.1083/jcb.200810029. J Cell Biol. 2009. PMID: 19414608 Free PMC article. - Nuclear pore complex function in Saccharomyces cerevisiae is influenced by glycosylation of the transmembrane nucleoporin Pom152p.
Belanger KD, Gupta A, MacDonald KM, Ott CM, Hodge CA, Cole CM, Davis LI. Belanger KD, et al. Genetics. 2005 Nov;171(3):935-47. doi: 10.1534/genetics.104.036319. Epub 2005 Aug 22. Genetics. 2005. PMID: 16118201 Free PMC article. - The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function.
Ho AK, Raczniak GA, Ives EB, Wente SR. Ho AK, et al. Mol Biol Cell. 1998 Feb;9(2):355-73. doi: 10.1091/mbc.9.2.355. Mol Biol Cell. 1998. PMID: 9450961 Free PMC article. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French. - Structure, dynamics and function of nuclear pore complexes.
D'Angelo MA, Hetzer MW. D'Angelo MA, et al. Trends Cell Biol. 2008 Oct;18(10):456-66. doi: 10.1016/j.tcb.2008.07.009. Epub 2008 Sep 9. Trends Cell Biol. 2008. PMID: 18786826 Free PMC article. Review.
Cited by
- The SESA network links duplication of the yeast centrosome with the protein translation machinery.
Sezen B, Seedorf M, Schiebel E. Sezen B, et al. Genes Dev. 2009 Jul 1;23(13):1559-70. doi: 10.1101/gad.524209. Genes Dev. 2009. PMID: 19571182 Free PMC article. - Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly.
Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, Antonin W. Vollmer B, et al. EMBO J. 2012 Oct 17;31(20):4072-84. doi: 10.1038/emboj.2012.256. Epub 2012 Sep 7. EMBO J. 2012. PMID: 22960634 Free PMC article. - Nuclear pore biogenesis into an intact nuclear envelope.
Doucet CM, Hetzer MW. Doucet CM, et al. Chromosoma. 2010 Oct;119(5):469-77. doi: 10.1007/s00412-010-0289-2. Epub 2010 Aug 19. Chromosoma. 2010. PMID: 20721671 Review. - The ESCRT-III complex is required for nuclear pore complex sequestration and regulates gamete replicative lifespan in budding yeast meiosis.
Koch BA, Staley E, Jin H, Yu HG. Koch BA, et al. Nucleus. 2020 Dec;11(1):219-236. doi: 10.1080/19491034.2020.1812872. Nucleus. 2020. PMID: 32893723 Free PMC article. - Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle.
Tamm T, Grallert A, Grossman EP, Alvarez-Tabares I, Stevens FE, Hagan IM. Tamm T, et al. J Cell Biol. 2011 Oct 31;195(3):467-84. doi: 10.1083/jcb.201106076. J Cell Biol. 2011. PMID: 22042620 Free PMC article.
References
- Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel and R. W. Wozniak, 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131: 1133–1148. - PMC - PubMed
- Allen, N. P., L. Huang, A. Burlingame and M. Rexach, 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276: 29268–29274. - PubMed
- Allen, T. D., J. M. Cronshaw, S. Bagley, E. Kiseleva and M. W. Goldberg, 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113: 1651–1659. - PubMed
- Antonin, W., and I. W. Mattaj, 2005. Nuclear pore complexes: round the bend? Nat. Cell Biol. 7: 10–12. - PubMed
- Antonin, W., C. Franz, U. Haselmann, C. Antony and I. W. Mattaj, 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell 17: 83–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases