The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH - PubMed (original) (raw)
The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH
Ivan B Lomakin et al. EMBO J. 2006.
Abstract
Eukaryotic initiation factor eIF1 and the functional C-terminal domain of prokaryotic initiation factor IF3 maintain the fidelity of initiation codon selection in eukaryotes and prokaryotes, respectively, and bind to the same regions of small ribosomal subunits, between the platform and initiator tRNA. Here we report that these nonhomologous factors can bind to the same regions of heterologous subunits and perform their functions in heterologous systems in a reciprocal manner, discriminating against the formation of initiation complexes containing codon-anticodon mismatches. We also show that like IF3, eIF1 can influence initiator tRNA selection, which occurs at the stage of ribosomal subunit joining after eIF5-induced hydrolysis of eIF2-bound GTP. The mechanisms of initiation codon and initiator tRNA selection in prokaryotes and eukaryotes are therefore unexpectedly conserved and likely involve related conformational changes induced in the small ribosomal subunit by factor binding. YciH, a prokaryotic eIF1 homologue, could perform some of IF3's functions, which justifies the possibility that YciH and eIF1 might have a common evolutionary origin as initiation factors, and that IF3 functionally replaced YciH in prokaryotes.
Figures
Figure 1
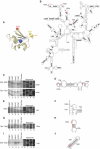
Binding of IF3, YciH and eIF1 to (A–C) 40S subunits and (D) 30S subunits. (A, B, D) Interaction of ribosomal subunits with T7-tag antibody agarose-immobilized eIF1, IF3, IF3-CTD or YciH (as indicated), and (B) in the presence of recombinant untagged eIF1 (as indicated) in in vitro binding assays. Ribosomal proteins and initiation factors were stained with Coomassie blue. Initiation factors are indicated by red arrows. (C) Presence of T7-tagged eIF1 and IF3 in ribosomal complexes isolated from sucrose density gradients (lanes 1 and 2), and eIF1 and IF3 markers (lanes 3 and 4) visualized by Western blotting.
Figure 2
Directed hydroxyl radical cleavage of 16S rRNA in 30S/eIF1 complexes from Fe(II) tethered to different positions on eIF1. (A) Ribbon diagram of the structured domain of eIF1. Colored spheres indicate the positions of cysteines introduced to tether Fe(II)-BABE. (B) Secondary structure of Escherichia coli 16S rRNA. Sites of directed hydroxyl radical cleavage are shown as red bars. (C, E, G, I) Primer extension analysis of directed hydroxyl radical cleavage of 16S rRNA (in helices 23, 44, 24 and 26, respectively) from Fe(II) tethered to positions on eIF1 as indicated. Cleavage sites are indicated to the left of each panel. Lanes marked ‘Cys-less' correspond to reaction mixtures that contained the cysteine-less eIF1 mutant. Lanes C, T, A and G depict 16S rRNA sequence generated from the same primer. (D, F, H, J) Elements of helices 23, 44, 24 and 26 of 16S rRNA with hydroxyl radical cleavage sites (red circles).
Figure 3
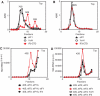
Activities of eIF1, IF3, IF3-CTD and YciH in promoting ribosomal dissociation and formation of 43S complexes. Dissociation of (A) 70S and (B) 80S ribosomes in the presence of factors as indicated. The optical density of ribosomal profiles was measured after centrifugation through 10–30% sucrose density gradients. The positions of ribosomes and ribosomal subunits are indicated. (C, D) 43S complex formation in reaction mixtures containing [35S]Met-tRNAiMet, 40S subunits and factors as indicated was assayed by centrifugation through 10–30% sucrose density gradients. Aliquots of gradient fractions were analyzed by scintillation counting. The position of 43S complexes is indicated. Fractions from upper parts of gradients were omitted for clarity.
Figure 4
Activities of eIF1, IF3, IF3-CTD and YciH in dissociating aberrant eukaryotic 48S complexes. (A) Sequences of 5′-UTRs of β-globin mRNA and (CAA)_n_-GUS mRNA derivatives with initiation codons in bold. (B–F) Toe-printing analysis of 48S complexes assembled on mRNAs as indicated. Reaction mixtures contained 40S subunits, Met-tRNAiMet and factors as indicated. The positions of toe-prints caused by assembled 48S complexes are shown on the left. Full-length cDNAs are labeled. (E) ‘Complex I' indicates the position of toe-prints caused by ribosomal complexes whose leading edge was 21–24 nt. from the 5′-end of β-globin mRNA. (B, C) Lanes marked C, T, A and G show cDNA sequences derived using the same primer as that for toe-printing.
Figure 5
Activities of eIF1, IF3 and YciH in initiation codon and initiator tRNA selection in prokaryotes. (A) The sequences of the 5′UTRs of SD-AUG and SD-AUU mRNAs. The Shine–Dalgarno sequence is bold and underlined; initiation codons are bold. (B–D) Toe-printing analysis of prokaryotic initiation complexes assembled on (B, D) SD-AUG mRNA and (C) SD-AUU mRNA. Reaction mixtures contained 30S subunits, IF1 and wild-type E.coli tRNAiMet or CCC-GGG mutant transcript tRNAiMet (Figure 6A) as well as IF3, eIF1 and YciH as indicated. Assembly reactions in (D) were carried out at 6 or 10 mM Mg2+ as indicated. Full-length cDNAs are labeled; cDNAs labeled ‘+16 nt. from AUG' or ‘+16 nt. from AUU' correspond to toe-prints caused by initiation complexes. Lanes marked G, A, T and C show cDNA sequences derived using the same primer as that for toe-printing.
Figure 6
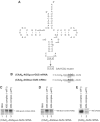
The activities of eIF1, IF3 and YciH in discriminating between wt and mutant forms of Met-tRNAiMet with mutations in the anticodon stem during 48S complex formation on (CAA)_n_-GUS mRNA. (A) The structure of wt tRNAiMet showing mutated nucleotides and the sequences of mutations in the anticodon stems of CCC-GGG and AGU-UCA mutant transcripts. (B) The sequence of 5′-UTR of (CAA)_n_-GUS mRNA showing the initiation codon in bold. (C–F) Toe-printing analysis of 48S complex formation on (CAA)_n_-GUS mRNA in reaction mixtures containing 40S subunits, factors, native eukaryotic Met-tRNAiMet, wt eukaryotic transcript Met-tRNAiMet, AGU-UCA or CCC-GGG mutant transcript Met-tRNAiMet, as indicated. Full-length cDNAs are labeled. The label ‘48S (GUS)' indicates the position of toe-prints caused by 48S complexes assembled on the GUS gene AUG. Lanes (C, T, A and G) show cDNA sequences derived using the same primer.
Figure 7
eIF1 does not discriminate against noncanonical complementary codon–anticodon base pairs. (A) Structure of wt tRNAiMet showing mutated nucleotides and the sequences of mutations in the anticodon loop of AGU-UCA mutant transcripts. (B) The sequence of the 5′-UTR of (CAA)_n_-AGGgood-GUS and (CAA)_n_-AGGbad-GUS mRNAs with initiation codons in bold. Context residues from −3 to +4 positions are underlined. (C–E) Toe-printing analysis of 48S complex formation on (CAA)_n_-AGGgood-GUS, (CAA)_n_-AGGbad-GUS and (CAA)_n_-GUS mRNAs in reaction mixtures containing 40S subunits, AGU-UCA mutant Met-tRNAiMet, eIFs 2, 3, 1A, 4B, 4B, 4F and eIF1, as indicated. The labels to the right of each panel show the position of toe-prints caused by 48S complexes assembled on AGG or AUG codons, as indicated.
Figure 8
Activities of eIF1, IF3, IF3-CTD and YciH in discriminating against initiation on the CrPV IGR IRES. (A, B) Toe-printing analysis of binding of 40S, 30S subunits, 80S and 70S ribosomes to this IRES in the presence of eIF1, IF3, IF3-CTD, YciH, and eEF1, eEF2 and Ala-tRNAAla as indicated. The 40S/80S-dependent toe-prints and toe-prints that appear following ribosomal translocation are indicated to the right. Full-length cDNAs are labeled. (C) Influence of IF3 on translation mediated by the CrPV IRES. In total, 0.2 μg of bicistronic RLuc-CrPV IRES-FLuc mRNA were translated for 60 min at 30°C in 15 μl of reticulocyte lysate that had been preincubated for 10 min with increasing amounts (0.2–1.5 μg) of IF3. Samples were analyzed by SDS–PAGE and autoradiography. (D) Structure of the CrPV IRES (Jan and Sarnow, 2002) showing the three pseudoknots, and toe-prints (black arrows) due to binding of 40S subunits (Wilson et al, 2000). mRNA triplets located in E, P and A sites are indicated.
Similar articles
- Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing.
Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Lomakin IB, et al. Genes Dev. 2003 Nov 15;17(22):2786-97. doi: 10.1101/gad.1141803. Epub 2003 Nov 4. Genes Dev. 2003. PMID: 14600024 Free PMC article. - Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection.
Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Valásek L, et al. Mol Cell Biol. 2004 Nov;24(21):9437-55. doi: 10.1128/MCB.24.21.9437-9455.2004. Mol Cell Biol. 2004. PMID: 15485912 Free PMC article. - Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP.
Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Unbehaun A, et al. Genes Dev. 2004 Dec 15;18(24):3078-93. doi: 10.1101/gad.1255704. Genes Dev. 2004. PMID: 15601822 Free PMC article. - The scanning mechanism of eukaryotic translation initiation.
Hinnebusch AG. Hinnebusch AG. Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review. - Molecular mechanisms of translation initiation in eukaryotes.
Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Pestova TV, et al. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7029-36. doi: 10.1073/pnas.111145798. Proc Natl Acad Sci U S A. 2001. PMID: 11416183 Free PMC article. Review.
Cited by
- Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex.
Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Pisarev AV, et al. Genes Dev. 2006 Mar 1;20(5):624-36. doi: 10.1101/gad.1397906. Genes Dev. 2006. PMID: 16510876 Free PMC article. - Migration of Small Ribosomal Subunits on the 5' Untranslated Regions of Capped Messenger RNA.
Shirokikh NE, Dutikova YS, Staroverova MA, Hannan RD, Preiss T. Shirokikh NE, et al. Int J Mol Sci. 2019 Sep 10;20(18):4464. doi: 10.3390/ijms20184464. Int J Mol Sci. 2019. PMID: 31510048 Free PMC article. - Identification and role of functionally important motifs in the 970 loop of Escherichia coli 16S ribosomal RNA.
Saraiya AA, Lamichhane TN, Chow CS, SantaLucia J Jr, Cunningham PR. Saraiya AA, et al. J Mol Biol. 2008 Feb 22;376(3):645-57. doi: 10.1016/j.jmb.2007.11.102. Epub 2007 Dec 7. J Mol Biol. 2008. PMID: 18177894 Free PMC article. - Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning.
Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Abaeva IS, et al. EMBO J. 2011 Jan 5;30(1):115-29. doi: 10.1038/emboj.2010.302. Epub 2010 Nov 26. EMBO J. 2011. PMID: 21113134 Free PMC article. - Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit.
Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hashem Y, et al. Nature. 2013 Nov 28;503(7477):539-43. doi: 10.1038/nature12658. Epub 2013 Nov 3. Nature. 2013. PMID: 24185006 Free PMC article.
References
- Cigan AM, Feng L, Donahue TF (1988) tRNAi(Met) functions in directing the scanning ribosome to the start site of translation. Science 242: 93–97 - PubMed
- Dallas A, Noller HF (2001) Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864 - PubMed
- Drabkin HJ, Helk B, RajBhandary UL (1993) The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J Biol Chem 268: 25221–25228 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI051340/AI/NIAID NIH HHS/United States
- R01 GM059660/GM/NIGMS NIH HHS/United States
- R01 AI51340/AI/NIAID NIH HHS/United States
- R01 GM59660/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous