The nuclear envelope: form and reformation - PubMed (original) (raw)
Review
The nuclear envelope: form and reformation
Amy J Prunuske et al. Curr Opin Cell Biol. 2006 Feb.
Abstract
The membrane system that encloses genomic DNA is referred to as the nuclear envelope. However, with emerging roles in signaling and gene expression, these membranes clearly serve as more than just a physical barrier separating the nucleus and cytoplasm. Recent progress in our understanding of nuclear envelope architecture and composition has also revealed an intriguing connection between constituents of the nuclear envelope and human disease, providing further impetus to decipher this cellular structure and the dramatic remodeling process it undergoes with each cell division.
Figures
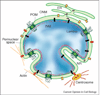
Figure 1
Schematic diagram of the nucleus, highlighting membrane domains of the nuclear envelope (NE) and associated structures. The membrane system of the nuclear envelope consists of the outer nuclear membrane (ONM), the inner nuclear membrane (INM) and the pore membrane (POM). The ONM is contiguous with the endoplasmic reticulum (ER). Portions of the NE extend into the nucleus forming the nucleoplasmic reticulum (NR). The INM contains many distinct proteins (black) that contact the underlying lamina and chromatin. The pore membrane houses integral membrane proteins of nuclear pore complexes (green). Some ONM proteins (yellow) are also present within the ER and others (red) preferentially localize to the ONM and are proposed to bridge INM proteins to such cytoplasmic structures as the centrosome and actin filaments. Finally, another category of protein (blue ovals) is able to diffuse within the perinuclear space and to interact with luminal domains of NE proteins.
Figure 2
Model of reformation of the nuclear envelope (NE) at the end of mitosis. Chromatin-associated RanGEF creates a gradient of RanGTP around DNA, which induces the localized release of nucleoporins (green balls) chaperoned at mitosis by importin β (orange). Importin α (red) also participates in nuclear formation and is, in part, membrane-associated. Some inner nuclear membrane (INM) proteins (grey), present on membrane, target to the chromatin during assembly. Formation of a closed NE requires incorporation of nuclear pore complexes into the fusing membrane. NE growth requires the addition of more membrane and pores as well as import through the nuclear pore complexes. Additional INM proteins (black), synthesized in the ER, target to the INM via the pore membrane (POM).
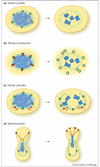
Figure 3
Mechanistic models of nuclear envelope (NE) disassembly. Key findings in various experimental systems are schematically depicted. Cells are not drawn to scale; it is notable that the oocyte is very large compared to a somatic cell, since size may impose specific constraints on the mechanics of NE breakdown. (a) In starfish oocytes, early alterations in permeability at the nuclear pore (green) have been observed, and correlate with an early phase of nuclear pore complex (NPC) disassembly. During the second phase of disassembly, larger holes in the NE are proposed to emanate from the site of disassembled pores. (b) In embryonic-like nuclei formed in vitro from Xenopus egg extract, nuclear pore proteins recruit the COPI complex (red) to the NE. Local concentration of this coatomer complex may then lead to vesiculation of the NE, as depicted, or to a non-conventional role for COPI. (c) In human tissue culture cells (somatic), microtubules originating from the centrosomes (yellow) connect to the NE via the microtubule motor dynein (black). Dynein-mediated movement is thought to then pull the NE toward the centrosomes, eventually causing a rupture at a distal region of the NE. (d) In Ustilago maydis, a basidiomycete, the NE is dragged from the mother cell to the bud by microtubules and dynein (black). There is an early increase in permeability, suggestive of pore remodeling, and then an obvious opening in the NE near the spindle pole body (yellow). Subsequently, the chromosomes enter the daughter cell where the spindle is formed, and the remnant of the NE collapses into the mother cell.
Similar articles
- Pushing the envelope: structure, function, and dynamics of the nuclear periphery.
Hetzer MW, Walther TC, Mattaj IW. Hetzer MW, et al. Annu Rev Cell Dev Biol. 2005;21:347-80. doi: 10.1146/annurev.cellbio.21.090704.151152. Annu Rev Cell Dev Biol. 2005. PMID: 16212499 Review. - The nuclear envelope: emerging roles in development and disease.
Wolfner MF, Wilson KL. Wolfner MF, et al. Cell Mol Life Sci. 2001 Nov;58(12-13):1737-40. doi: 10.1007/PL00000812. Cell Mol Life Sci. 2001. PMID: 11766874 Free PMC article. - Composition of the plant nuclear envelope: theme and variations.
Meier I. Meier I. J Exp Bot. 2007;58(1):27-34. doi: 10.1093/jxb/erl009. Epub 2006 Jun 27. J Exp Bot. 2007. PMID: 16804054 Review. - Nuclear envelope proteins and their role in nuclear positioning and replication.
Graumann K, Runions J, Evans DE. Graumann K, et al. Biochem Soc Trans. 2010 Jun;38(3):741-6. doi: 10.1042/BST0380741. Biochem Soc Trans. 2010. PMID: 20491659 Review. - Nuclear envelope rupture drives genome instability in cancer.
Lim S, Quinton RJ, Ganem NJ. Lim S, et al. Mol Biol Cell. 2016 Nov 1;27(21):3210-3213. doi: 10.1091/mbc.E16-02-0098. Mol Biol Cell. 2016. PMID: 27799497 Free PMC article.
Cited by
- Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly.
Gorjánácz M, Klerkx EP, Galy V, Santarella R, López-Iglesias C, Askjaer P, Mattaj IW. Gorjánácz M, et al. EMBO J. 2007 Jan 10;26(1):132-43. doi: 10.1038/sj.emboj.7601470. Epub 2006 Dec 14. EMBO J. 2007. PMID: 17170708 Free PMC article. - Nuclear size rectification: A potential new therapeutic approach to reduce metastasis in cancer.
Schirmer EC, Latonen L, Tollis S. Schirmer EC, et al. Front Cell Dev Biol. 2022 Oct 10;10:1022723. doi: 10.3389/fcell.2022.1022723. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36299481 Free PMC article. Review. - Systems biology meets chromatin function: a report on the Fourth Elmau Conference on Nuclear Organization.
Lavelle C, Sigal A. Lavelle C, et al. Chromosome Res. 2007;15(2):247-56. doi: 10.1007/s10577-006-1118-6. Epub 2007 Jan 26. Chromosome Res. 2007. PMID: 17279452 - Coordinating postmitotic nuclear pore complex assembly with abscission timing.
Mackay DR, Ullman KS. Mackay DR, et al. Nucleus. 2011 Jul-Aug;2(4):283-8. doi: 10.4161/nucl.2.4.16189. Epub 2011 Jul 1. Nucleus. 2011. PMID: 21941107 Free PMC article. - A Yeast Mitotic Tale for the Nucleus and the Vacuoles to Embrace.
Santana-Sosa S, Matos-Perdomo E, Ayra-Plasencia J, Machín F. Santana-Sosa S, et al. Int J Mol Sci. 2023 Jun 6;24(12):9829. doi: 10.3390/ijms24129829. Int J Mol Sci. 2023. PMID: 37372977 Free PMC article. Review.
References
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. A subtractive proteomic approach is used to identify 67 previously uncharacterized nuclear envelope proteins. This adds to an earlier proteomic approach where 19 uncharacterized nuclear envelope proteins were identified [1].
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. - PubMed
- Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. - PubMed
- Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, Theodoropoulos PA, Singh PB, Georgatos SD. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. 2004;279:25567–25573. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources