Translational control in positive strand RNA plant viruses - PubMed (original) (raw)
Review
Translational control in positive strand RNA plant viruses
Theo W Dreher et al. Virology. 2006.
Abstract
The great variety of genome organizations means that most plant positive strand viral RNAs differ from the standard 5'-cap/3'-poly(A) structure of eukaryotic mRNAs. The cap and poly(A) tail recruit initiation factors that support the formation of a closed loop mRNA conformation, the state in which translation initiation is most efficient. We review the diverse array of cis-acting sequences present in viral mRNAs that compensate for the absence of a cap, poly(A) tail, or both. We also discuss the cis-acting sequences that control translation strategies that both amplify the coding potential of a genome and regulate the accumulations of viral gene products. Such strategies include leaky scanning initiation of translation of overlapping open reading frames, stop codon readthrough, and ribosomal frameshifting. Finally, future directions for research on the translation of plant positive strand viruses are discussed.
Figures
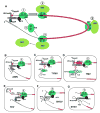
Fig. 1
The closed loop scheme for initiation of translation: thematic variations used by plant positive strand RNA viruses. Panel A shows in simplified terms (not all factors are shown; not to scale) the circularized format in which efficiently translated cellular mRNAs are believed to exist. The key responsible cis-acting features are the 5_′m7GpppN cap (purple dot) and 3_′_-poly(A) tail, which synergistically enhance expression. Bridging interactions through eIF4E or eIF-iso4E (the cap-binding proteins), eIF4G or eIF-iso4G, and the poly(A) binding protein (PABP) bring the 5_′_- and 3_′_-termini into close proximity, and the interactions are mutually stabilizing. The first stage of translation initiation, the recruitment of the 40S small ribosome subunit to the 5_′_-end, depends on simultaneous interaction of eIF3 (a complex of multiple proteins) with eIF4G or eIF-iso4G and the 40S subunit. This step is depicted in step (1) and is followed by ribosome scanning (arrow) along the 5_′_-UTR (black line) in search of the AUG initiation codon. In response to base-pairing with the initiator tRNA (not shown) located in the ribosomal P site, the 60S large ribosome subunit joins (step 2) to initiate the elongation phase of translation. During peptide elongation (step 3), the codons in the ORF (thick red line) are read by tRNAs entering the ribosomal A site, until a termination codon, such as UAA, is reached (step 4), triggering termination of protein synthesis and subunit dissociation and release (step 5). Because of the closed loop format, ribosomes are near the 5_′_-end upon termination, facilitating new initiation. The boxed diagrams (B–G) illustrate variations of the initial 40S subunit recruitment step for viral mRNAs that lack a cap, poly(A) tail, or both, and which are discussed in the text. Red question marks indicate unknown or uncertain details. AMV, TYMV, and TMVare examples of viruses whose RNAs have a cap but no poly(A) tail. For AMV, a coat protein (CP) dimer binds to the 3_′_-terminal region and to eIF4G/iso4G. For TYMV RNA, aminoacylation (indicated by Val in the diagram) of the 3_′_-terminal tRNA-like structure (TLS) is needed for full 3_′_-translational enhancement, and it has been postulated that eEF1A binding is involved in closed loop formation. TMV RNA also has a 3_′ TLS capable of aminoacylation (His) and eEF1A binding, but 3_′_-translational enhancement relies on an upstream pseudoknot (UPSK). Intriguingly, this feature can also bind eEF1A, which may be involved in closed loop formation, apparently in a way that predominates over a TYMV-like interaction involving the TLS. TEV RNA has a poly(A) tail, but no 5_′_-cap. The 5_′_-end is covalently linked to VPg, which is not needed for translation but does interact with eIF4E and eIF-iso4E; it is not known whether this interaction influences translation. 40S ribosome subunits are recruited to the 5_′_-UTR through an IRES element whose function requires eIF4G and that may involve direct base-pairing to 18S ribosomal RNA. BYDV and STNV RNAs lack both canonical terminal elements and possess translational enhancer elements (BTE, TED) in an internal position (not at the 3_′_-terminus) of the 3_′_-UTR. These elements recruit translation initiation factors that are normally recruited to the 5_′_-end by the cap. In BYDV RNA, ribosome delivery to the 5_′_-end is accomplished through direct RNA base-pairing between elements in the 5_′_- and 3_′_-UTRs, while in STNV RNA, base-pairing between the 5_′_-UTR and rRNA may be involved. See text for details.
Fig. 2
Expanded expression repertoire resulting from leaky scanning and translational recoding. Panel A depicts in simple terms the coding content of a standard mRNA, which directs protein synthesis between an initiation codon, typically the 5_′_-most AUG (black diamond), and a termination codon (red hexagon). The encoded protein is indicated by the thick line below the RNA. (B) In many viral RNAs, more than one initiation site can be used. This can occur by leaky scanning when the 5_′_-most initiation codon is weakly recognized by ribosomes because it is in a weak context, as occurs with pyrimidines (Y) in the −3 and/or +4 positions (as indicated) or if initiation occurs at a non-AUG codon (not shown). When the initiation sites are in different reading frames, two proteins of unrelated sequence are made (as shown); when the initiation sites are in-frame (not shown), the encoded proteins are identical except for the presence of an N-terminal extension on one of the proteins. The product of the downstream ORF is generally less efficiently expressed, indicated by the thinner line. (C) In viral RNAs such as TMV RNA, a suppressible termination codon (cross-hatched hexagon) is embedded in an ORF. In conjunction with the downstream recoding signal CARYYA (R = purine; Y = pyrimidine), a small proportion of ribosomes avoids termination, permitting the synthesis of an elongated version of the upstream protein. (D) Frameshifting (typically −1) occurs in mRNAs, which have a pair of recoding signals: a ‘‘slippery’’ heptanucleotide (XXxNNNZ, where X and N can be any base and Z is any base except G) and a feature such as a pseudoknot that is thought to induce ribosome pausing. The encoded products represent translation of the entire upstream ORF and a longer chimeric protein derived from the different ORFs upstream and downstream of the frameshift point.
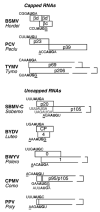
Fig. 3
Examples of viral genes translated by leaky scanning. Only the ORFs involved in leaky scanning are shown. The genus to which each virus belongs is listed below the virus acronym. Gaps in box outline indicate ORFs that are not shown to scale. Initiation codon contexts are shown above (first ORF) or below (second ORF) the translation start site. Bases at −3 and +4 positions relative to the start codon (bold AUG), that fit the optimal context (G at +4, A at −3) are underlined. In all cases, the second ORF start codon is in a better initiation context than the upstream AUGs. The p20 ORF of SBMV-C contains two AUG codons in weak contexts that do not act as initiation codons (gray sequences below p20 ORF) (Sivakumaran and Hacker, 1998). The second start codons of CPMVand PPVare in the same reading frame as the first, yielding an N-terminally truncated protein. The CPMV ORF has an in-frame AUG (gray sequence) 12 nt downstream of the second start codon that can act as a start codon in artificial contexts, but is unlikely to function under usual conditions (Holness et al., 1989). See text for additional explanation and references.
Similar articles
- Translation of Plant RNA Viruses.
Geng G, Wang D, Liu Z, Wang Y, Zhu M, Cao X, Yu C, Yuan X. Geng G, et al. Viruses. 2021 Dec 13;13(12):2499. doi: 10.3390/v13122499. Viruses. 2021. PMID: 34960768 Free PMC article. Review. - 3' cap-independent translation enhancers of plant viruses.
Simon AE, Miller WA. Simon AE, et al. Annu Rev Microbiol. 2013;67:21-42. doi: 10.1146/annurev-micro-092412-155609. Epub 2013 May 13. Annu Rev Microbiol. 2013. PMID: 23682606 Free PMC article. Review. - 3' Cap-independent translation enhancers of positive-strand RNA plant viruses.
Nicholson BL, White KA. Nicholson BL, et al. Curr Opin Virol. 2011 Nov;1(5):373-80. doi: 10.1016/j.coviro.2011.10.002. Epub 2011 Oct 24. Curr Opin Virol. 2011. PMID: 22440838 Review. - In vitro translation of plant viral RNA.
Browning KS, Mayberry L. Browning KS, et al. Curr Protoc Microbiol. 2006 Jun;Chapter 16:16K.1.1-16K.1.13. doi: 10.1002/9780471729259.mc16k01s01. Curr Protoc Microbiol. 2006. PMID: 18770587 - Short 5' Untranslated Region Enables Optimal Translation of Plant Virus Tricistronic RNA via Leaky Scanning.
Fujimoto Y, Keima T, Hashimoto M, Hagiwara-Komoda Y, Hosoe N, Nishida S, Nijo T, Oshima K, Verchot J, Namba S, Yamaji Y. Fujimoto Y, et al. J Virol. 2022 Apr 13;96(7):e0214421. doi: 10.1128/jvi.02144-21. Epub 2022 Mar 9. J Virol. 2022. PMID: 35262378 Free PMC article.
Cited by
- Advances in Understanding the Mechanism of Cap-Independent Cucurbit Aphid-Borne Yellows Virus Protein Synthesis.
Truniger V, Pechar GS, Aranda MA. Truniger V, et al. Int J Mol Sci. 2023 Dec 18;24(24):17598. doi: 10.3390/ijms242417598. Int J Mol Sci. 2023. PMID: 38139425 Free PMC article. - Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence.
Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Dabrowski M, et al. RNA Biol. 2015;12(9):950-8. doi: 10.1080/15476286.2015.1068497. RNA Biol. 2015. PMID: 26176195 Free PMC article. Review. - Oscillating kissing stem-loop interactions mediate 5' scanning-dependent translation by a viral 3'-cap-independent translation element.
Rakotondrafara AM, Polacek C, Harris E, Miller WA. Rakotondrafara AM, et al. RNA. 2006 Oct;12(10):1893-906. doi: 10.1261/rna.115606. Epub 2006 Aug 18. RNA. 2006. PMID: 16921068 Free PMC article. - A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells.
Castorena KM, Weeks SA, Stapleford KA, Cadwallader AM, Miller DJ. Castorena KM, et al. J Virol. 2007 Aug;81(16):8412-20. doi: 10.1128/JVI.00189-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522196 Free PMC article.
References
- Aranda M, Maule A. Virus-induced host gene shutoff in animals and plants. Virology. 1998;243:261–267. - PubMed
- Baranov PV, Gesteland RF, Atkins JF. Recording: translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
- Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous