E47 is required for V(D)J recombinase activity in common lymphoid progenitors - PubMed (original) (raw)
Comparative Study
E47 is required for V(D)J recombinase activity in common lymphoid progenitors
Lisa Borghesi et al. J Exp Med. 2005.
Abstract
Common lymphoid progenitors (CLPs) are the first bone marrow precursors in which V(D)J recombinase activity is up-regulated. Here, we show that loss of the transcription factor E47 produces a reduced CLP population that lacks V(D)J recombinase activity and D-J(H) rearrangements in vivo. Apart from a profound arrest before the pro-B cell stage, other downstream lymphoid progeny of CLPs are still intact in these mice albeit at reduced numbers. In contrast to the inhibition of recombinase activity in early B lineage precursors in E47-deficient animals, loss of either E47 or its cis-acting target Erag (enhancer of rag transcription) has little effect on recombinase activity in thymic T lineage precursors. Taken together, this work defines a role for E47 in regulating lineage progression at the CLP stage in vivo and describes the first transcription factor required for lineage-specific recombinase activity.
Figures
Figure 1.
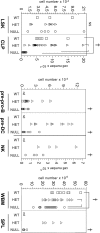
The B lineage defect in E47-null mice. (A) Bone marrow from E47-heterozygous or E47-null mice crossed to the H2-SVEX (SB110) background was stained with antibodies to detect CD19− CD24lo pre-pro–B cells and CD19+ CD24hi pro–/pre–B cells within the B220+ AA4.1+ subset. Contaminating populations expressing DX5, Ly6C, IgM, or CD4 were excluded from the analysis. Pre-pro–B cells were then analyzed for VEX expression. (B) Pre-pro–B cells were examined for flt3 expression. (C) B lineage progenitors were identified according to an alternative phenotypic model in which CD19− CD24lo pre-pro–B cells and CD19+ CD24hi pro–B cells are identified within the B220+ CD43+ subset. As in A, cells bearing DX5, Ly6C, IgM, or CD4 are excluded from the analysis. B cell progenitor populations were subsequently analyzed for VEX expression. The data are representative of three to five independent experiments.
Figure 2.
Quantification of lymphoid populations in E47-deficient animals. E47 wild-type, E47-heterozygous, or E47-null mice were evaluated for the presence of the indicated hematopoietic subsets. Within each hematopoietic population, each symbol represents an independent animal. See Results for markers used to identify each population. ND, not done; NS, not significant. †, P < 0.05.
Figure 3.
Effects of E_rag_ and E47 on recombinase activity in T lineage progenitors. (A) Thymocytes from E47 wild-type, E47-null, E_rag_ wild-type, or E_rag_-null mice crossed to the H2-SVEX (SB110) background were stained with antibodies to CD3, CD4, and CD8 to identify immature T cell subsets or lineage markers (CD4, CD8α, CD3ɛ, γδ-TCR, NK1.1, Mac-1, Gr-1, and B220), CD44, and CD25 to identify the earliest T lineage progenitors. TN (TN1–TN4) populations are: CD44+ CD25− lin−, CD44+ CD25+ lin−, CD44− CD25+ lin−, and CD44− CD25− lin−, respectively. These populations were then analyzed for VEX expression. The data are representative of at least two independent experiments.
Figure 4.
Recombinase activity in DCs and NK cells. (A) Bone marrow DC precursors (AA4.1+ B220+ CD4+ CD24lo) or (B) splenic NK cells (NK1.1+ CD122+ CD19− CD3− CD4− CD8−) from E47-heterozygous or E47-null mice crossed to the H2-SVEX (SB110) background were examined for VEX expression. The data are representative of two to four independent experiments.
Figure 5.
Recombinase activity in CLPs. Bone marrow from E47-heterozygous or E47-null mice was stained with antibodies to detect (A) LSKs (lin− Sca-1hi c-Kithi where lineage markers are NK1.1, CD11b, Gr-1, TER-119, CD3, CD19, and B220) or (B) CLPs (IL7R+ lin− AA4.1+ Sca-1lo). Results from three independent experiments are shown to provide perspective on the scope of the CLP defect across multiple E47-deficient animals. Gates were drawn with respect to the relevant controls within each experiment. (C) Bone marrow CLPs were then examined for c-Kit and flt3 expression. (D) Bone marrow CLPs in E47-heterozygous or E47-null mice were identified using the following phenotypic scheme that does not rely on the AA4.1 marker: IL7R+ lin− Sca-1lo c-Kitlo. AA4.1 expression is provided for comparison to B. The data are representative of 3–10 independent experiments.
Figure 6.
Apoptosis and proliferation in multipotent progenitors. (A) Bone marrow CLPs and pre-pro–B cells from E47-heterozygous or E47-knockout mice were stained with annexin V and DAPI to identify live, apoptotic, and dead cells. As a positive control for annexin V detection, thymocytes from B6 mice were cultured with 1 μM dexamethasone for 5 h to induce apoptosis. (B) Bone marrow from E47-heterozygous or E47-deficient mice injected with BrdU (shaded histogram) was then stained for surface markers to identify CLPs or pre-pro–B cells followed by intracellular staining with anti-BrdU antibodies. Background staining (open histogram) was determined by injecting E47-heterozygous mice with PBS followed by the identical staining procedures. The data are representative of two to three independent experiments.
Figure 7.
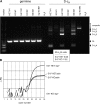
Recombination at endogenous IgH loci. (A) Bone marrow CLPs as in Fig. 5 D were examined for D-JH rearrangement status at the endogenous IgH locus by PCR. Germline (GL) DNA and rearrangements of D to J1, J2, J3, or J4 is indicated by arrowheads. Data from two independent sorts is shown (E47 heterozygote no. 1 and E47 knockout no. 1, 100 CLPs each; E47 heterozygote no. 2 and E47 knockout no. 2, 150 CLPs each). Rearrangement status of whole bone marrow from wild-type B6 mice and from rag2 knockout mice is provided for comparison. Inset depicts the frequency of CLPs that have detectable D-JH rearrangements as determined by quantitative single cell PCR. (B) rag1 expression was quantified by real-time RT-PCR using 350 cell equivalents of cDNA from CLPs. Expression of rag1 and the reference gene β-actin are shown for equal numbers of CLPs from E47-heterozygous and E47 knockout animals. ΔRN, the fluorescence intensity. Identical results were obtained using cells from at least two independent sorts.
Similar articles
- B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors.
Borghesi L, Hsu LY, Miller JP, Anderson M, Herzenberg L, Herzenberg L, Schlissel MS, Allman D, Gerstein RM. Borghesi L, et al. J Exp Med. 2004 Feb 16;199(4):491-502. doi: 10.1084/jem.20031800. Epub 2004 Feb 9. J Exp Med. 2004. PMID: 14769852 Free PMC article. - Developmental separation of V(D)J recombinase expression and initiation of IgH recombination in B lineage progenitors in vivo.
Borghesi L, Gerstein RM. Borghesi L, et al. J Exp Med. 2004 Feb 16;199(4):483-9. doi: 10.1084/jem.20031802. Epub 2004 Feb 9. J Exp Med. 2004. PMID: 14769853 Free PMC article. - Induction of sterile transcription from the kappa L chain gene locus in V(D)J recombinase-deficient progenitor B cells.
Grawunder U, Rolink A, Melchers F. Grawunder U, et al. Int Immunol. 1995 Dec;7(12):1915-25. doi: 10.1093/intimm/7.12.1915. Int Immunol. 1995. PMID: 8746561 - Bone marrow microenvironmental changes in aged mice compromise V(D)J recombinase activity and B cell generation.
Labrie JE 3rd, Borghesi L, Gerstein RM. Labrie JE 3rd, et al. Semin Immunol. 2005 Oct;17(5):347-55. doi: 10.1016/j.smim.2005.05.012. Semin Immunol. 2005. PMID: 15963731 Review. - Transcriptional regulation in early B cell development.
Fuxa M, Skok JA. Fuxa M, et al. Curr Opin Immunol. 2007 Apr;19(2):129-36. doi: 10.1016/j.coi.2007.02.002. Epub 2007 Feb 9. Curr Opin Immunol. 2007. PMID: 17292598 Review.
Cited by
- The Function of E2A in B-Cell Development.
Miyazaki M, Miyazaki K. Miyazaki M, et al. Adv Exp Med Biol. 2024;1459:97-113. doi: 10.1007/978-3-031-62731-6_5. Adv Exp Med Biol. 2024. PMID: 39017841 Review. - In Vitro Human Haematopoietic Stem Cell Expansion and Differentiation.
Bozhilov YK, Hsu I, Brown EJ, Wilkinson AC. Bozhilov YK, et al. Cells. 2023 Mar 14;12(6):896. doi: 10.3390/cells12060896. Cells. 2023. PMID: 36980237 Free PMC article. Review. - Receptor editing constrains development of phosphatidyl choline-specific B cells in VH12-transgenic mice.
Worth AN, Palmer VL, Schabla NM, Perry GA, Fraser-Philbin AN, Swanson PC. Worth AN, et al. Cell Rep. 2022 Jun 14;39(11):110899. doi: 10.1016/j.celrep.2022.110899. Cell Rep. 2022. PMID: 35705027 Free PMC article. - Helix-Loop-Helix Proteins in Adaptive Immune Development.
Aubrey M, Warburg ZJ, Murre C. Aubrey M, et al. Front Immunol. 2022 May 12;13:881656. doi: 10.3389/fimmu.2022.881656. eCollection 2022. Front Immunol. 2022. PMID: 35634342 Free PMC article. Review. - The influence of space and time on the establishment of B cell identity.
Elsaid R, Yang J, Cumano A. Elsaid R, et al. Biomed J. 2019 Aug;42(4):209-217. doi: 10.1016/j.bj.2019.07.008. Epub 2019 Sep 19. Biomed J. 2019. PMID: 31627863 Free PMC article. Review.
References
- Igarashi, H., S.C. Gregory, T. Yokota, N. Sakaguchi, and P.W. Kincade. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 17:117–130. - PubMed
- Schwarz, B.A., and A. Bhandoola. 2004. Circulating hematopoietic progenitors with T lineage potential. Nat. Immunol. 5:953–960. - PubMed
- Bain, G., E.C. Robanus Maandag, H.P. te Riele, A.J. Feeney, A. Sheehy, M. Schlissel, S.A. Shinton, R.R. Hardy, and C. Murre. 1997. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 6:145–154. - PubMed
- Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell. 79:875–884. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous