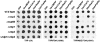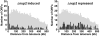The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control - PubMed (original) (raw)
The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control
David J Dilworth et al. J Cell Biol. 2005.
Abstract
Nuclear pore complexes (NPCs) govern macromolecular transport between the nucleus and cytoplasm and serve as key positional markers within the nucleus. Several protein components of yeast NPCs have been implicated in the epigenetic control of gene expression. Among these, Nup2p is unique as it transiently associates with NPCs and, when artificially tethered to DNA, can prevent the spread of transcriptional activation or repression between flanking genes, a function termed boundary activity. To understand this function of Nup2p, we investigated the interactions of Nup2p with other proteins and with DNA using immunopurifications coupled with mass spectrometry and microarray analyses. These data combined with functional assays of boundary activity and epigenetic variegation suggest that Nup2p and the Ran guanylyl-nucleotide exchange factor, Prp20p, interact at specific chromatin regions and enable the NPC to play an active role in chromatin organization by facilitating the transition of chromatin between activity states.
Figures
Figure 1.
Prp20p and Nup2p interact with chromatin remodeling factors. (a) Prp20p is nucleosome associated. (Left) Prp20p-prA was immunopurified from yeast whole-cell lysates and abundant copurifying proteins were identified by MS analysis of gel slices. Components of the histone octamer, H2A, H2B, H3, and H4, were present as well as the linker histone, H1 and Ran/Gsp1p. The presence of Mgm101p is not specific (see text). (Right) Prp20p-prA eluates contain DNA. Eluates were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Prp20p-prA associated DNA is ∼600 bp in length due to chromatin shearing during the cell lysis procedures. (b) The interaction between Nup2p and the Prp20p–nucleosome complex can be reconstituted in vitro. (Top) Bacterially expressed and purified Prp20p was incubated with glutathione resin coated with GST or GST-Nup2p. Unbound and bound proteins were resolved by SDS-PAGE and visualized with Coomassie blue. (Middle) The Prp20p–nucleosome complex was bound to IgG-coated magnetic beads and then incubated with bacterially expressed and purified GST-Nup2p-CFP, GST-CFP, or GST-Nup2p. Immunoblotting for GST revealed that although GST alone could not bind to the Prp20p–nucleosome complex, GST chimeras containing Nup2p bound efficiently (asterisks). Arrows indicate nonspecific immunoreactive proteins. (Bottom) Glutathione resin coated with GST-Nup2p, but not GST alone, was able to capture Prp20p-prA from yeast extracts. (c) Interactions between the NPC and chromatin. Proteins present at Coomassie blue–detectable levels in Nup2p, Nup60p, and/or Prp20p immunopurifications are connected by solid black lines. Other known physical and yeast two-hybrid interactions are shown by dotted and dashed lines, respectively (Dingwall et al., 1995; Rexach and Blobel, 1995; Denning et al., 2001; Feuerbach et al., 2002). High coverage tandem MS (MS/MS) of immunopurification eluates from Nup2p, Prp20p, and Nup60p,as well as Kap95p and Nup49p (as controls) was also performed. The inset list (Eluate MS/MS) shows proteins present exclusively in Nup2p eluates (top) or Prp20p eluates (bottom) and those present in both eluates (middle). (d) Nucleosomes associated with Prp20p and Htz1p possess unique acetylation patterns suggestive of boundary chromatin. The acetylation levels of residues K5, K8, K12, and K16 of histone H4 were quantified by mass spectrometry for global (white), Prp20p-associated (dark gray), and Htz1p-associated (light gray) nucleosomes.
Figure 2.
The boundary trap assay confirms the links between the NPC and chromatin-bound Prp20p. The ability of Nup2p to bind to the NPC through Nup60p is required for BA. The boundary trap strain, KIY54, and isogenic Δnup2, Δnup60, Δmlp1, Δmlp2, and Δmlp1/Δmlp2 derivatives expressing plasmids encoding the Gal4 DNA binding domain alone, Gbd (pGBC11) or fused to the COOH-terminal portion of the Drosophila BEAF protein, Gbd-BEAF (pGBC11-BEAF-C), a GFP-tagged portion of Cse1p, Gbd-Cse1p (pGBC11-CSE1[474–960]-GFP), full-length Nup2p, Gbd-Nup2p (pGBC12-NUP2[1–720]) or GFP-tagged Prp20p, Gbd-Prp20p-GFP (pGBC12-PRP20-GFP) were serially spotted onto CSM-TRP (T), CSM-TRP+FOA (TF) and CSM-TRP-ADE+FOA (TAF) to assess boundary function. Cells lacking Nup60p were defective in their ability to silence the URA3 reporter, indicated by a reduced viability on media containing 5-FOA. This phenotype was also shared by two independently isolated double-mutant strains lacking both of the Mlp proteins. BA is indicated by growth on media lacking adenine and containing 5-FOA (TAF). A plasmid encoding only Gbd failed to elicit BA and the positive control fusion, Gbd-BEAF, exhibited BA in all genotypes tested. The BA of Gbd-Cse1p was dependent on Nup2p (Ishii et al., 2002), and BA of both Gbd-Cse1p and Gbd-Nup2p was absent in Δnup60 mutants. Double-mutant Δmlp1/Δmlp2 strains exhibited reduced BA. Gdb-Prp20p possesses BA in wild-type and single Mlp mutant strains at frequencies comparable to transport factors (Ishii et al., 2002). Deletion of NUP2 or NUP60 or both MLP1 and MLP2 resulted in dramatically reduced Gbd-Prp20p BA.
Figure 3.
Genes exhibiting aberrant expression in cells lacking Nup2p map to distinct chromosome regions. For the top 5% of significant Δnup2 induced or repressed ORFs, the distance from each ORF to the nearest telomere was determined. These distances were grouped into 10-kb bins and plotted as a function of telomeric distance. These plots reveal an enrichment of _Δnup2_-induced ORFs at subtelomeric regions, as 25% of induced ORFs reside within 20 kb of a chromosome end. Only 1% of significantly repressed ORFs were within this distance. The shaded histograms indicate the distribution of telomeric distances for all ORFs plotted at 1/8 scale on the y-axis. Statistical comparison of the _Δnup2_-induced and repressed distributions to the profile for all ORFs using a two-sample Kolmogorov-Smirnov test confirms that the induced profile is unique (P = 0.0000386), but the repressed profile is not (P = 0.287).
Figure 4.
DNA regions bound by Prp20p and Nup2p enrich near telomeres and lie in close proximity to ORFs induced in cells lacking Nup2p. (a) Histograms of minimal telomeric distance for the top 5% of significantly enriched intergenic regions bound by Prp20p and Nup2p reveal a telomeric enrichment similar to that observed for ORFs induced in cells lacking Nup2p (see Fig. 3). The shaded histograms represent the distribution of all intergenic regions shown at 1/4 scale on the y-axis. The Prp20p and Nup2p profiles are significantly distinct from the distribution of all intergenic regions (P < 0.000001 and P = 0.000367, respectively). In contrast, the profile of the transcription factor Oaf1p displayed no significant enrichment relative to all intergenic regions (P = 0.368; not depicted). (b) Chromosomal proximity of transcriptionally induced ORFs in Δnup2 cells and ChIP-CHIP enriched intergenic regions. The distance between each enriched intergenic region and the nearest _Δnup2_-induced ORF was determined for Nup2p, Prp20p, Oaf1p, and 10 randomized datasets. Intergenic regions bound by Nup2p and Prp20p are found much closer to ORFs induced in cells lacking Nup2p, relative to the Oaf1p or randomized datasets, as evidenced by the high number of Nup2p and Prp20p enriched intergenic regions found within 10 kb of Δnup2 ORFs. The Kolmogorov-Smirnov test reveals significant differences between the Oaf1p profile and those obtained with Prp20p and Nup2p (P = 0.0282 and P = 0.00102, respectively).
Figure 5.
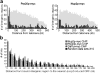
Genetic interactions support links between NUP 2, NUP60, PRP20, and HTZ1. Growth rate analysis of Δhtz1, Δnup2, Δnup60, Δnup_53, and prp20-7 single-mutant and relevant double-mutant strains at 23, 30, and 37°C. Double-mutant prp20-7 Δ_htz1, Δhtz1 Δ_nup2_, Δhtz1 Δ_nup60_, prp20-7 Δ_nup2_, and prp20-7 Δ_nup60_ strains all exhibited more severe growth defects than those detected in their parental strains, whereas double-mutant combinations involving deletion of NUP53 revealed no genetic interactions.
Figure 6.
Loss of Nup2p or Gsp2p results in subtelomeric gene silencing defects that are distinct from those observed for strains lacking Nup60p or Htz1p. (a) The expression status of a telomerically encoded α2 reporter gene was assayed in single cells by monitoring the response of cells to the α-factor. Cells that do not express α2 (OFF) respond to α-factor arrest in G1 and shmoo; cells expressing α2 (ON) continue to bud and divide. The initial OFF proportion was determined by scoring cells after a 4-h treatment with α-factor. (b) Determination of OFF maintenance ratio. α2 OFF maintenance was assayed by monitoring α-factor arrested cells over time. Cells that have switched to the ON state give rise to microcolonies, whereas stably arrested cells do not divide. (c) The wild-type normalized initial OFF ratios (y-axis) and OFF maintenance ratios (x-axis) were plotted for each strain (error bars indicate the SD for three independent experiments). Strains lacking Nup2p or Gsp2p exhibited very similar phenotypes (marginally increased initial OFF ratios [y ≥ 1], and poor OFF state maintenance [x < 1]). Cells lacking Nup60p exhibited a steady-state defect in the establishment of the OFF state and an inability to maintain the OFF state (x and y < 1), whereas cells lacking Htz1p showed the opposite phenotype (x and y > 1).
Figure 7.
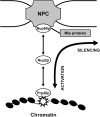
Dynamic model of NPC-mediated BA. Boundaries (star), marked by Prp20p, are proposed to be mobile but spatially restricted within the nucleus due to their transient Nup2p-dependent association with NPCs. The complexation of DNA with the NPC represents an unstable reaction intermediate from which the DNA can either enter the perinuclear silencing region through Nup60p or detach from the NPC, free to enter the nuclear interior.
Similar articles
- The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex.
Denning D, Mykytka B, Allen NP, Huang L, Al Burlingame, Rexach M. Denning D, et al. J Cell Biol. 2001 Sep 3;154(5):937-50. doi: 10.1083/jcb.200101007. J Cell Biol. 2001. PMID: 11535617 Free PMC article. - Chromatin boundaries in budding yeast: the nuclear pore connection.
Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Ishii K, et al. Cell. 2002 May 31;109(5):551-62. doi: 10.1016/s0092-8674(02)00756-0. Cell. 2002. PMID: 12062099 - Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes.
Lapetina DL, Ptak C, Roesner UK, Wozniak RW. Lapetina DL, et al. J Cell Biol. 2017 Oct 2;216(10):3145-3159. doi: 10.1083/jcb.201609049. Epub 2017 Sep 7. J Cell Biol. 2017. PMID: 28883038 Free PMC article. - The multifunctional nuclear pore complex: a platform for controlling gene expression.
Ptak C, Aitchison JD, Wozniak RW. Ptak C, et al. Curr Opin Cell Biol. 2014 Jun;28:46-53. doi: 10.1016/j.ceb.2014.02.001. Epub 2014 Mar 22. Curr Opin Cell Biol. 2014. PMID: 24657998 Free PMC article. Review. - The Nuclear Pore Complex in Cell Type-Specific Chromatin Structure and Gene Regulation.
Sun J, Shi Y, Yildirim E. Sun J, et al. Trends Genet. 2019 Aug;35(8):579-588. doi: 10.1016/j.tig.2019.05.006. Epub 2019 Jun 15. Trends Genet. 2019. PMID: 31213386 Review.
Cited by
- Interchromosomal clustering of active genes at the nuclear pore complex.
Brickner DG, Brickner JH. Brickner DG, et al. Nucleus. 2012 Nov-Dec;3(6):487-92. doi: 10.4161/nucl.22663. Epub 2012 Oct 25. Nucleus. 2012. PMID: 23099887 Free PMC article. - The nuclear pore complex: bridging nuclear transport and gene regulation.
Strambio-De-Castillia C, Niepel M, Rout MP. Strambio-De-Castillia C, et al. Nat Rev Mol Cell Biol. 2010 Jul;11(7):490-501. doi: 10.1038/nrm2928. Nat Rev Mol Cell Biol. 2010. PMID: 20571586 Review. - Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization.
Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Titus LC, et al. Mol Biol Cell. 2010 Mar 15;21(6):1072-87. doi: 10.1091/mbc.e09-07-0615. Epub 2010 Jan 28. Mol Biol Cell. 2010. PMID: 20110349 Free PMC article. - A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.
Bonnet A, Chaput C, Palmic N, Palancade B, Lesage P. Bonnet A, et al. PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article. - Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes.
Dieppois G, Iglesias N, Stutz F. Dieppois G, et al. Mol Cell Biol. 2006 Nov;26(21):7858-70. doi: 10.1128/MCB.00870-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954382 Free PMC article.
References
- Aebi, M., M.W. Clark, U. Vijayraghavan, and J. Abelson. 1990. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol. Gen. Genet. 224:72–80. - PubMed
- Aitchison, J.D., M.P. Rout, M. Marelli, G. Blobel, and R.W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. - PMC - PubMed
- Akhtar, N., H. Hagan, J.E. Lopilato, and A.H. Corbett. 2001. Functional analysis of the yeast Ran exchange factor Prp20p: in vivo evidence for the RanGTP gradient model. Mol. Genet. Genomics. 265:851–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR022220/RR/NCRR NIH HHS/United States
- P41 RR000862/RR/NCRR NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- RR00862/RR/NCRR NIH HHS/United States
- GM066496/GM/NIGMS NIH HHS/United States
- F32 GM066496/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous