Microbial pattern recognition receptors mediate M-cell uptake of a gram-negative bacterium - PubMed (original) (raw)
Comparative Study
Microbial pattern recognition receptors mediate M-cell uptake of a gram-negative bacterium
Peter Tyrer et al. Infect Immun. 2006 Jan.
Abstract
The receptors involved in the sampling of particulate microbial antigens by the gut are largely unknown. Here we demonstrate for the first time in an in vitro M-cell model and in situ in isolated murine intestinal segments that the receptors TLR-4, PAF-R, and alpha5beta1 integrin are all involved in mediating bacterial uptake associated with transcytosis. The pattern of expression of TLR-4 and alpha5beta1 integrin differed between M cells and enterocytes. There was increased apical expression of TLR-4 in M-cell cultures, and it was present on the apical surface of murine M cells but not enterocytes in situ. In contrast, PAF-R was expressed equally by both cell types in vitro and was abundantly expressed throughout the intestinal epithelium. Inhibition of TLR-4 and PAF-R, but not TLR-2, reduced gram-negative bacterial uptake by both cell types, whereas inhibition of the apically expressed alpha5beta1 integrin significantly reduced the ability of M cells to translocate bacteria. Hence, the involvement of each receptor was dependent not only on differences in the level of receptor expression but the cellular localization. Using bacteria that had mutations that affected the bacterial lipooligosaccharide structure indicated that the oligosaccharide moiety was important in bacterial uptake. Taken together, the data suggest that pathogen-associated molecular pattern interactions with pattern recognition receptors are key factors in M-cell recognition of intestinal antigens for mucosal immune priming.
Figures
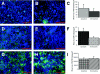
FIG. 1.
Expression of PRRs by in vitro M-cell cocultures and Caco-2 monocultures as measured by immunofluorescence microscopy and image analysis with MetaMorph software. (A to C) Expression of TLR-2. TLR-2 was probed with anti-TLR-2 antibody and antibody conjugated Alexa 488 (green). (A) Expression of TLR-2 in M-cell cocultures; (B) expression of TLR-2 in Caco-2 monocultures; (C) image analysis of expression of TLR-2. Image analysis demonstrates that there was no difference in expression between the culture systems (P = 0.217). (D to F) Expression of TLR-4. TLR-4 was probed with anti-TLR-4 antibody and antibody conjugated Alexa 488 (green). (D) Expression of TLR-4 by M-cell cocultures; (E) expression of TLR-4 by Caco-2 monocultures; (F) image analysis of expression of TLR-4. Image analysis demonstrates that TLR-4 was significantly upregulated in the M-cell cocultures (P = 0.043). (G to I) Expression of PAF-R. PAF-R was detected with anti-PAF-R antibody and antibody-conjugated Alexa 488 conjugate (green). (G) Expression of PAF-R by M-cell cocultures; (H) expression of PAF-R by Caco-2 monocultures; (I) image analysis of PAF-R expression. Image analysis demonstrated that PAF-R was expressed equally by both cell culture systems (_P_=0.432). In each photomicrograph, nuclei are counterstained blue with DAPI. Size bar, 100 μm. The image analysis data are expressed as the mean± the standard error of the mean (SEM).
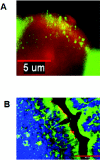
FIG. 2.
Expression of PRRs by murine M cells and enterocytes in an intestinal segment as shown by immunofluorescence microscopy. M cells were identified by the M-cell-specific Ulex europeaus (I) TRITC conjugate (red). TLR-4 and PAF-R were probed with anti-TLR-4 (A) and anti-PAF-R (B) and stained with antibody-conjugated Alexa 488 (green for both TLR-4 and PAF-R). (A) TLR-4 (green) was expressed on the apical surfaces of murine M cells (red) and intracellularly in adjacent follicle-associated enterocytes, as seen in this representative longitudinal section. Size bar, 5 μm. (B) PAF-R (green) was expressed ubiquitously throughout the murine intestine, including M cells (red). The section is counterstained blue with DAPI. Size bar,50 μm.
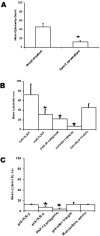
FIG. 3.
NTHi translocation by M-cell cocultures and Caco-2 monocultures. Translocation of fluorescently labeled NTHi 289 from the lower chamber (apical surface) to the upper chamber (basal surface) was measured by flow cytometry analysis of the culture media. (A) M-cell cocultures translocated more NTHi 289 than Caco-2 monocultures (P = 0.009). (B) In the M-cell cocultures, specific blocking of TLR-4 (P = 0.046), PAF-R (P = 0.004), and α5β1 integrin (P < 0.001) inhibited transcytosis of killed NTHi. (C) In the Caco-2 monocultures, blocking of TLR-4 (P = 0.017) and PAF-R (P < 0.001) inhibited translocation of NTHi 289 by Caco-2 cells. The data are expressed as means ± the SEM, and the “*” indicates a significant difference from the control with the P values indicated above.
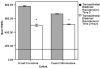
FIG. 4.
Effects of killed NTHi translocation on transepithelial electrical resistance in M-cell cocultures and Caco-2 monocultures. The TEERs of the M-cell cocultures and Caco-2 monocultures were determined by an EVOM meter on addition of killed NTHi to the apical chambers and after 2 h of incubation. Introduction of the NTHi to the cell cultures did result in a significant decrease in the TEER in both culture systems (P < 0.001) with the decrease being slightly greater in the M cell cocultures. The absolute levels of TEERs did not drop sufficiently to compromise the epithelial barrier function that occurs when the TEER drops to 150 Ω · cm2 (indicated by the line across the graph). The data are expressed as means ± the SEM, and the “*” indicates a significant difference from the control with the P values indicated above.
FIG. 5.
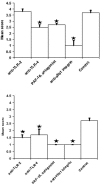
Blocking PRRs with specific receptor antagonists inhibits translocation of NTHi in situ in mouse intestinal segments. (A and B) Specific blocking of TLR-4 (P = 0.004), PAF-R (P = 0.013) and α5β1 integrin (P < 0.001) inhibited uptake of NTHi 289 into Peyer's patches (A) and TLR-2 (P = 0.003), TLR-4 (P < 0.001), PAF-R (P < 0.001), and α5β1 integrin (P < 0.001) (B) inhibited uptake of NTHi 289 into the intestinal villus. The data are expressed as means ± the SEM, and the “*” indicates a significant difference from control with the P values indicated above.
Similar articles
- Receptor mediated targeting of M-cells.
Tyrer PC, Ruth Foxwell A, Kyd JM, Otczyk DC, Cripps AW. Tyrer PC, et al. Vaccine. 2007 Apr 20;25(16):3204-9. doi: 10.1016/j.vaccine.2007.01.028. Epub 2007 Jan 17. Vaccine. 2007. PMID: 17276559 - Application of a mouse ligated Peyer’s patch intestinal loop assay to evaluate bacterial uptake by M cells.
Fukuda S, Hase K, Ohno H. Fukuda S, et al. J Vis Exp. 2011 Dec 17;(58):3225. doi: 10.3791/3225. J Vis Exp. 2011. PMID: 22215009 Free PMC article. - Peptidoglycan recognition protein expression in mouse Peyer's Patch follicle associated epithelium suggests functional specialization.
Lo D, Tynan W, Dickerson J, Mendy J, Chang HW, Scharf M, Byrne D, Brayden D, Higgins L, Evans C, O'Mahony DJ. Lo D, et al. Cell Immunol. 2003 Jul;224(1):8-16. doi: 10.1016/s0008-8749(03)00155-2. Cell Immunol. 2003. PMID: 14572796 - Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium.
Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Mabbott NA, et al. Mucosal Immunol. 2013 Jul;6(4):666-77. doi: 10.1038/mi.2013.30. Epub 2013 May 22. Mucosal Immunol. 2013. PMID: 23695511 Free PMC article. Review. - Intestinal M cells.
Ohno H. Ohno H. J Biochem. 2016 Feb;159(2):151-60. doi: 10.1093/jb/mvv121. Epub 2015 Dec 3. J Biochem. 2016. PMID: 26634447 Free PMC article. Review.
Cited by
- Hyaluronic Acid in the Intestinal Tract: Influence of Structure, Rheology, and Mucoadhesion on the Intestinal Uptake in Rats.
Barbosa de Souza A, Vinícius Chaud M, Francine Alves T, Ferreira de Souza J, Andrade Santana MH. Barbosa de Souza A, et al. Biomolecules. 2020 Oct 8;10(10):1422. doi: 10.3390/biom10101422. Biomolecules. 2020. PMID: 33050089 Free PMC article. - The SRC family tyrosine kinase HCK and the ETS family transcription factors SPIB and EHF regulate transcytosis across a human follicle-associated epithelium model.
Asai T, Morrison SL. Asai T, et al. J Biol Chem. 2013 Apr 12;288(15):10395-405. doi: 10.1074/jbc.M112.437475. Epub 2013 Feb 25. J Biol Chem. 2013. PMID: 23439650 Free PMC article. - The role of epithelial integrin receptors in recognition of pulmonary pathogens.
Ulanova M, Gravelle S, Barnes R. Ulanova M, et al. J Innate Immun. 2009;1(1):4-17. doi: 10.1159/000141865. Epub 2008 Jun 25. J Innate Immun. 2009. PMID: 20375562 Free PMC article. Review. - Identification of scavenger receptor B1 as the airway microfold cell receptor for Mycobacterium tuberculosis.
Khan HS, Nair VR, Ruhl CR, Alvarez-Arguedas S, Galvan Rendiz JL, Franco LH, Huang L, Shaul PW, Kim J, Xie Y, Mitchell RB, Shiloh MU. Khan HS, et al. Elife. 2020 Mar 5;9:e52551. doi: 10.7554/eLife.52551. Elife. 2020. PMID: 32134383 Free PMC article. - Nasal immunity to staphylococcal toxic shock is controlled by the nasopharynx-associated lymphoid tissue.
Fernandez S, Cisney ED, Hall SI, Ulrich RG. Fernandez S, et al. Clin Vaccine Immunol. 2011 Apr;18(4):667-75. doi: 10.1128/CVI.00477-10. Epub 2011 Feb 16. Clin Vaccine Immunol. 2011. PMID: 21325486 Free PMC article.
References
- Abreu, M. T., P. Vora, E. Faure, L. S. Thomas, E. T. Arnold, and M. Arditi. 2001. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J. Immunol. 167:1609-1616. - PubMed
- Akashi, S., S. Saitoh, Y. Wakabayashi, T. Kikuchi, N. Takamura, Y. Nagai, Y. Kusumoto, K. Fukase, S. Kusumoto, Y. Adachi, A. Kosugi, and K. Miyake. 2003. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. J. Exp. Med. 198:1035-1042. - PMC - PubMed
- Foxwell, A. R., A. W. Cripps, and K. B. Dear. 2003. Haemophilus influenzae oral vaccination against acute bronchitis. Cochrane Database Syst. Rev:CD001958. - PubMed
- Foxwell, A. R., J. M. Kyd, and A. W. Cripps. 1998. Kinetics of inflammatory cytokines in the clearance of nontypeable Haemophilus influenzae from the lung. Immunol. Cell Biol. 76:556-559. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases