The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells - PubMed (original) (raw)
The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells
Marc S Weinberg et al. RNA. 2006 Feb.
Abstract
To determine mechanistically how siRNAs mediate transcriptional gene silencing (TGS) in human cells, we have measured histone methylation at targeted promoters, the dependency on active transcription, and whether or not both strands of the siRNA are required for siRNA-mediated TGS. We report here that siRNA treatment increases both H3K9 and H3K27 methylation of the targeted EF1A promoter and that this increase is dependent on nuclear specific delivery of the siRNA. We also find that TGS can be directed by the antisense strand alone, and requires active transcription by RNA polymerase II in human cells as evidenced by sensitivity to alpha-amanatin. The observation of antisense strand-specific siRNA-mediated TGS of EF1A was substantiated by targeting the U3 region of the HIV-1 LTR/promoter. Furthermore, we show that the antisense strand of siRNA EF52 associates with the transiently expressed Flag-tagged DNMT3A, the targeted EF1A promoter, and trimethylated H3K27. The observations reported here implicate a functional link between siRNA-mediated targeting of genomic regions (promoters), RNA Pol II function, histone methylation, and DNMT3A and support a paradigm in which the antisense strands of siRNAs alone can direct sequence-specific transcriptional gene silencing in human cells.
Figures
FIGURE 1.
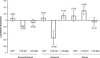
siRNA-mediated histone methylation and the requirement for RNA polymerase II function. (A) MPG-transfected EF52 siRNA induces histone methylation. Histone 3 lysine 9 (H3K9) dimethylation and histone 3 lysine 27 (H3K27) trimethylation were determined from 293T cells transfected with EF52 or the control CCR5 siRNAs (10 nM) using the nuclear-specific amphipathic peptide MPG (Morris et al. 1997). Forty-eight hours post-transfection, ChIP assays were performed specifically for the EF1A promoter (Morris et al. 2004a). Results represent a minimum of two independent experiments with the range shown. (B) Nuclear-specific delivery is required for histone methylation. MPG and Lipofectamine 2000 transfection reagents were used to transfect EF52-Cy3+ siRNAs into 293T cells. Forty-eight hours following transfection, cultures were collected and a ChIP assay was performed. (C) H3K9 dimethylation can spread 720 bp downstream of the targeted EF1A promoter. H3K9 dimethylation was measured by quantitative PCR either 196 or 720 bp downstream from the targeted EF52 promoter. siRNAs EF52 and CCR5 (10 nM) were transfected with MPG into 293T cells, and ChIP assays were performed 48 h later. Two independent experiments with the range are shown. (D) Treatment of 293T cells with α-amanatin (0.05 μg/mL) 24 h following transfection with siRNA EF52 reduces H3K9 methylation to levels comparable to the no-antibody controls. Results from three independent experiments are shown with the standard deviations.
FIGURE 2.
siRNA interactions with DNMT3A. (A) Western blot analysis following biotin/avidin pull-down shows that antisense and dsRNA EF52 associate with DNMT3A but not DNMT1. Flag-tagged DNMT1 and DNMT3A transfected 293T cell lysates were generated and incubated with a total of 500 nM biotin-labeled sense (S), anti-sense (AS), or double-stranded (S/AS) siRNA, and the control dsRNA without a biotin label (−). Immunostaining for the Western blot was performed with an anti-Flag antibody. (B) Western blot analysis showing comparable expression levels of both Flag-tagged DNMT1 and DNMT3A in the transfected 293T cell lysates.
FIGURE 3.
LTR-247 sense/antisense and antisense alone are capable of targeting the U3 region of the HIV-1 LTR and inhibiting Tat-mediated expression of fire fly luciferase in 1G5 cells. Results are from four independent experiments measured 24 h post-transfection with log relative expression and standard error of the means shown. Values represent luciferase expression relative to the experimental mean.
FIGURE 4.
Biotin-labeled antisense siRNA EF52 (AS+Biotin) coprecipitates with trimethylated H3K27, Flag-tagged DNMT3A, and the targeted EF1A promoter, while the sense EF52 (S+Biotin) does not. Results are representative of two independent experiments.
Similar articles
- Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana.
Sasaki T, Lee TF, Liao WW, Naumann U, Liao JL, Eun C, Huang YY, Fu JL, Chen PY, Meyers BC, Matzke AJ, Matzke M. Sasaki T, et al. Plant J. 2014 Jul;79(1):127-38. doi: 10.1111/tpj.12545. Epub 2014 Jun 13. Plant J. 2014. PMID: 24798377 - Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro.
Lim HG, Suzuki K, Cooper DA, Kelleher AD. Lim HG, et al. Mol Ther. 2008 Mar;16(3):565-70. doi: 10.1038/sj.mt.6300380. Epub 2008 Jan 29. Mol Ther. 2008. PMID: 18227841 - Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells.
Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Kim DH, et al. Nat Struct Mol Biol. 2006 Sep;13(9):793-7. doi: 10.1038/nsmb1142. Epub 2006 Aug 27. Nat Struct Mol Biol. 2006. PMID: 16936726 - RNA-mediated transcriptional gene silencing in human cells.
Morris KV. Morris KV. Curr Top Microbiol Immunol. 2008;320:211-24. doi: 10.1007/978-3-540-75157-1_10. Curr Top Microbiol Immunol. 2008. PMID: 18268846 Review. - Transcriptional gene silencing by short interfering RNAs.
Kawasaki H, Taira K. Kawasaki H, et al. Curr Opin Mol Ther. 2005 Apr;7(2):125-31. Curr Opin Mol Ther. 2005. PMID: 15844619 Review.
Cited by
- Promoter-associated RNAs and promoter-targeted RNAs.
Yan BX, Ma JX. Yan BX, et al. Cell Mol Life Sci. 2012 Sep;69(17):2833-42. doi: 10.1007/s00018-012-0953-1. Epub 2012 Mar 14. Cell Mol Life Sci. 2012. PMID: 22415323 Free PMC article. Review. - Roles of the Core Components of the Mammalian miRISC in Chromatin Biology.
La Rocca G, Cavalieri V. La Rocca G, et al. Genes (Basel). 2022 Feb 24;13(3):414. doi: 10.3390/genes13030414. Genes (Basel). 2022. PMID: 35327968 Free PMC article. Review. - H3K4me2 and WDR5 enriched chromatin interacting long non-coding RNAs maintain transcriptionally competent chromatin at divergent transcriptional units.
Subhash S, Mishra K, Akhade VS, Kanduri M, Mondal T, Kanduri C. Subhash S, et al. Nucleic Acids Res. 2018 Oct 12;46(18):9384-9400. doi: 10.1093/nar/gky635. Nucleic Acids Res. 2018. PMID: 30010961 Free PMC article. - miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA.
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. Hansen TB, et al. EMBO J. 2011 Sep 30;30(21):4414-22. doi: 10.1038/emboj.2011.359. EMBO J. 2011. PMID: 21964070 Free PMC article. - Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells.
Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Napoli S, et al. EMBO J. 2009 Jun 17;28(12):1708-19. doi: 10.1038/emboj.2009.139. Epub 2009 May 21. EMBO J. 2009. PMID: 19461583 Free PMC article.
References
- Aguilar-Cordova, E., Chinen, J., Donehower, L., Lewis, D.E., and Belmont, J.W. 1994. A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res. Hum. Retroviruses 10: 295–301. - PubMed
- Castanotto, D., Tommasi, S., Li, M., Li, H., Yanow, S., Pfeifer, G.P., and Rossi, J.J. 2005. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol. Ther. 12: 179–183. - PubMed
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. 2004. RNA silencing genes control de novo DNA methylation. Science 303: 1336. - PubMed
- Danzer, J.R. and Wallrath, L.L. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131: 3571–3580. - PubMed
- Datta, J., Ghoshal, K., Sharma, S.M., Tajima, S., and Jacob, S.T. 2003. Biochemical fractionation reveals association of DNA methyltransferase (Dnmt) 3b with Dnmt1 and that of Dnmt 3a with a histone H3 methyltransferase and Hdac1. J. Cell Biochem. 88: 855–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5P30 CA33572/CA/NCI NIH HHS/United States
- R01 HL083473/HL/NHLBI NIH HHS/United States
- R01 HL07470/HL/NHLBI NIH HHS/United States
- R01 HL83473/HL/NHLBI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases