Topology of three-way junctions in folded RNAs - PubMed (original) (raw)
Topology of three-way junctions in folded RNAs
Aurélie Lescoute et al. RNA. 2006 Jan.
Abstract
The three-way junctions contained in X-ray structures of folded RNAs have been compiled and analyzed. Three-way junctions with two helices approximately coaxially stacked can be divided into three main families depending on the relative lengths of the segments linking the three Watson-Crick helices. Each family has topological characteristics with some conservation in the non-Watson-Crick pairs within the linking segments as well as in the types of contacts between the segments and the helices. The most populated family presents tertiary interactions between two helices as well as extensive shallow/minor groove contacts between a linking segment and the third helix. On the basis of the lengths of the linking segments, some guidelines could be deduced for choosing a topology for a three-way junction on the basis of a secondary structure. Examples and prediction bas'ed on those rules are discussed.
Figures
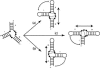
FIGURE 1.
Example of a hypothetical three-way junction. The Watson-Crick paired helices are indicated by the ladders. Depending on the structure of the internal junctions, two of the three helices may stack coaxially. In the example shown, helices II and III stack coaxially. Arbitrary non-Watson-Crick contacts are shown as dotted lines. This coaxial stack is maintained whatever the precise secondary structure (whether two loops or only one or none are capped, or whether the closing loops are internal loops in longer helices). In case (a), potential tertiary contacts would occur between loops I and II; in case (b), the contacts would occur potentially between loop I and helix II (or an internal loop within helix II); and in case (c) the contacts would occur potentially between loop II and helix I (or an internal loop within helix I).
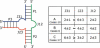
FIGURE 2.
The nomenclature used for the three-way junctions with the average numbers of nucleotides in each junction in the table at the right. The number of instances in each of the three families is also indicated.
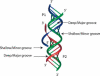
FIGURE 3.
Coaxial stack of two helices with single strands entering or leaving the helices. The deep/major and shallow/minor grooves are indicated. The 3′-end strand leaving helix P2 faces the deep/major groove of helix P1. The 5′-end strand entering helix P1 faces the shallow/minor groove of helix P2. One example can be seen in the structure of tRNA, in which the strand leaving the anti-codon hairpin (equivalent to P2) faces the deep/major groove of the dihydrouridine helix (equivalent to P1). Group I introns contain the case depicted at the junction between P4 and P6 (Michel et al. 1990; Adams et al. 2004; Guo et al. 2004; Golden et al. 2005; Woodson 2005).
FIGURE 4.
Schematic drawings of the three observed families, A, B, and C, in the analyzed three-way junctions. The drawings at the right are based on real structures. In family A, the third helix can adopt various angles with respect to the coaxially stacked helices.
FIGURE 5.
The three-way junctions belonging to family A. Ten three-way rRNAs junctions belong to family A. The name of the junction depends on the RNA to which it belongs and on the numbering of the three helices that are anchored to the junction. For a typical junction, the three-dimensional stereo view (DeLano Scientific,
) is shown on the right and the secondary structure with secondary and tertiary interactions is represented on the left (Yang et al. 2003). For the other junctions, only the secondary structure diagrams with the symbols for the tertiary contacts are shown. The symbols used are according to the Leontis and Westhof (2001) nomenclature.
FIGURE 6.
A consensus for the family A three-way junction. In family A, the junction J12 includes 0–5 nt; J23, 3–15 nt; and J31, 1–4 nt. Nucleotides at position 3, 4, or 5 of J23 make tertiary interactions with the shallow groove of the first one or the first two Watson-Crick base pairs of P2. The closing base pair of P2 is always in the trans orientation.
FIGURE 7.
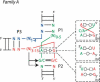
The three-way junctions belonging to family B. Same legend as for Figure 5 ▶.
FIGURE 8.
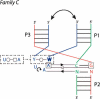
The three-way junctions belonging to family C. Ten cases come from the rRNA structures, and the other seven are from different RNA structures. Same legend as for Figure 5 ▶.
FIGURE 9.
A consensus for the family C three-way junction. In some cases, junction strand J31 folds into a tri-loop closed by a trans base pair, often a trans Watson-Crick. The nucleotide in 3′ is in the syn conformation (bold) and the free loop nucleotides at positions 2 and 3, often adenines, make Sugar-Edge–Sugar-Edge interactions with nucleotides of base pairs at positions 1 and 2 of helix P2. In almost all the observed cases, there are tertiary interactions between helices P1 and P3 (double arrow).
FIGURE 10.
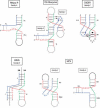
Some applications to noncrystallized three-way junctions. The secondary structures of the unknown RNAs have been represented in the fold corresponding to the proposed family. In the case of HCV, as two junction strands have no free nucleotides, two possibilities of stacking are allowed so the junction could belong to either family C or family A.
Similar articles
- Analysis of four-way junctions in RNA structures.
Laing C, Schlick T. Laing C, et al. J Mol Biol. 2009 Jul 17;390(3):547-59. doi: 10.1016/j.jmb.2009.04.084. Epub 2009 May 13. J Mol Biol. 2009. PMID: 19445952 Free PMC article. - Isostericity and tautomerism of base pairs in nucleic acids.
Westhof E. Westhof E. FEBS Lett. 2014 Aug 1;588(15):2464-9. doi: 10.1016/j.febslet.2014.06.031. Epub 2014 Jun 17. FEBS Lett. 2014. PMID: 24950426 Review. - Specificity of RNA-RNA helix recognition.
Battle DJ, Doudna JA. Battle DJ, et al. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11676-81. doi: 10.1073/pnas.182221799. Epub 2002 Aug 20. Proc Natl Acad Sci U S A. 2002. PMID: 12189204 Free PMC article. - Helix capping in RNA structure.
Lee JC, Gutell RR. Lee JC, et al. PLoS One. 2014 Apr 1;9(4):e93664. doi: 10.1371/journal.pone.0093664. eCollection 2014. PLoS One. 2014. PMID: 24691270 Free PMC article. - RNA structure and dynamics: a base pairing perspective.
Halder S, Bhattacharyya D. Halder S, et al. Prog Biophys Mol Biol. 2013 Nov;113(2):264-83. doi: 10.1016/j.pbiomolbio.2013.07.003. Epub 2013 Jul 23. Prog Biophys Mol Biol. 2013. PMID: 23891726 Review.
Cited by
- Long Intergenic Non-Coding RNAs: Novel Drivers of Human Lymphocyte Differentiation.
Panzeri I, Rossetti G, Abrignani S, Pagani M. Panzeri I, et al. Front Immunol. 2015 Apr 15;6:175. doi: 10.3389/fimmu.2015.00175. eCollection 2015. Front Immunol. 2015. PMID: 25926836 Free PMC article. Review. - Simultaneous prediction of RNA secondary structure and helix coaxial stacking.
Shareghi P, Wang Y, Malmberg R, Cai L. Shareghi P, et al. BMC Genomics. 2012 Jun 11;13 Suppl 3(Suppl 3):S7. doi: 10.1186/1471-2164-13-S3-S7. BMC Genomics. 2012. PMID: 22759616 Free PMC article. - A ribosome-binding, 3' translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction.
Gao F, Kasprzak W, Stupina VA, Shapiro BA, Simon AE. Gao F, et al. J Virol. 2012 Sep;86(18):9828-42. doi: 10.1128/JVI.00677-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761367 Free PMC article. - Naturally occurring three-way junctions can be repurposed as genetically encoded RNA-based sensors.
Moon JD, Wu J, Dey SK, Litke JL, Li X, Kim H, Jaffrey SR. Moon JD, et al. Cell Chem Biol. 2021 Nov 18;28(11):1569-1580.e4. doi: 10.1016/j.chembiol.2021.04.022. Epub 2021 May 18. Cell Chem Biol. 2021. PMID: 34010626 Free PMC article. - Three-dimensional structure of a flavivirus dumbbell RNA reveals molecular details of an RNA regulator of replication.
Akiyama BM, Graham ME, O Donoghue Z, Beckham JD, Kieft JS. Akiyama BM, et al. Nucleic Acids Res. 2021 Jul 9;49(12):7122-7138. doi: 10.1093/nar/gkab462. Nucleic Acids Res. 2021. PMID: 34133732 Free PMC article.
References
- Agalarov, S.C., Sridhar Prasad, G., Funke, P.M., Stout, C.D., and Williamson, J.R. 2000. Structure of the S15, S6, S18-rRNA complex: Assembly of the 30S ribosome central domain. Science 288: 107–113. - PubMed
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. - PubMed
- Batey, R.T., Rambo, R.P., and Doudna, J.A. 1999. Tertiary motifs in RNA structure and folding. Angew Chem. Int. Ed. Engl. 38: 2326–2343. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources