NOPdb: Nucleolar Proteome Database - PubMed (original) (raw)
NOPdb: Nucleolar Proteome Database
Anthony Kar Lun Leung et al. Nucleic Acids Res. 2006.
Abstract
The Nucleolar Proteome Database (NOPdb) archives data on >700 proteins that were identified by multiple mass spectrometry (MS) analyses from highly purified preparations of human nucleoli, the most prominent nuclear organelle. Each protein entry is annotated with information about its corresponding gene, its domain structures and relevant protein homologues across species, as well as documenting its MS identification history including all the peptides sequenced by tandem MS/MS. Moreover, data showing the quantitative changes in the relative levels of approximately 500 nucleolar proteins are compared at different timepoints upon transcriptional inhibition. Correlating changes in protein abundance at multiple timepoints, highlighted by visualization means in the NOPdb, provides clues regarding the potential interactions and relationships between nucleolar proteins and thereby suggests putative functions for factors within the 30% of the proteome which comprises novel/uncharacterized proteins. The NOPdb (http://www.lamondlab.com/NOPdb) is searchable by either gene names, nucleotide or protein sequences, Gene Ontology terms or motifs, or by limiting the range for isoelectric points and/or molecular weights and links to other databases (e.g. LocusLink, OMIM and PubMed).
Figures
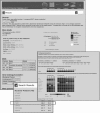
Figure 1
Snapshots of the NOPdb (
). The database was searched against molecular weights between 65 and 70 kDa and here we show an overview page for the PES1 protein (pescadillo) documenting its motif distribution, its GO annotations, its identification history by multiple MS analysis and its quantitative data from SILAC analyses. Proteins of similar kinetic profiles based on correlation coefficient are identified for future investigation. The kinetic profiles are ranked according to the Pearson's correlation coefficients for the log value of the peak intensities of multiple peptides at a particular timepoint normalized to the respective peak intensities at zero timepoints.
Similar articles
- NOPdb: Nucleolar Proteome Database--2008 update.
Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. Ahmad Y, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D181-4. doi: 10.1093/nar/gkn804. Epub 2008 Nov 4. Nucleic Acids Res. 2009. PMID: 18984612 Free PMC article. - The Nuclear Protein Database (NPD): sub-nuclear localisation and functional annotation of the nuclear proteome.
Dellaire G, Farrall R, Bickmore WA. Dellaire G, et al. Nucleic Acids Res. 2003 Jan 1;31(1):328-30. doi: 10.1093/nar/gkg018. Nucleic Acids Res. 2003. PMID: 12520015 Free PMC article. - Arabidopsis nucleolar protein database (AtNoPDB).
Brown JW, Shaw PJ, Shaw P, Marshall DF. Brown JW, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D633-6. doi: 10.1093/nar/gki052. Nucleic Acids Res. 2005. PMID: 15608277 Free PMC article. - Bioinformatic analysis of the nucleolus.
Leung AK, Andersen JS, Mann M, Lamond AI. Leung AK, et al. Biochem J. 2003 Dec 15;376(Pt 3):553-69. doi: 10.1042/BJ20031169. Biochem J. 2003. PMID: 14531731 Free PMC article. Review. - Deciphering the human nucleolar proteome.
Couté Y, Burgess JA, Diaz JJ, Chichester C, Lisacek F, Greco A, Sanchez JC. Couté Y, et al. Mass Spectrom Rev. 2006 Mar-Apr;25(2):215-34. doi: 10.1002/mas.20067. Mass Spectrom Rev. 2006. PMID: 16211575 Review.
Cited by
- Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA.
Boyne JR, Whitehouse A. Boyne JR, et al. Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15190-5. doi: 10.1073/pnas.0604890103. Epub 2006 Sep 27. Proc Natl Acad Sci U S A. 2006. PMID: 17005724 Free PMC article. - The proteins of intra-nuclear bodies: a data-driven analysis of sequence, interaction and expression.
Mohamad N, Bodén M. Mohamad N, et al. BMC Syst Biol. 2010 Apr 13;4:44. doi: 10.1186/1752-0509-4-44. BMC Syst Biol. 2010. PMID: 20388198 Free PMC article. - Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation.
Fu D, Collins K. Fu D, et al. Mol Cell. 2007 Dec 14;28(5):773-85. doi: 10.1016/j.molcel.2007.09.023. Mol Cell. 2007. PMID: 18082603 Free PMC article. - The estrogen and c-Myc target gene HSPC111 is over-expressed in breast cancer and associated with poor patient outcome.
Butt AJ, Sergio CM, Inman CK, Anderson LR, McNeil CM, Russell AJ, Nousch M, Preiss T, Biankin AV, Sutherland RL, Musgrove EA. Butt AJ, et al. Breast Cancer Res. 2008;10(2):R28. doi: 10.1186/bcr1985. Epub 2008 Mar 29. Breast Cancer Res. 2008. PMID: 18373870 Free PMC article. - Whole-genome screening identifies proteins localized to distinct nuclear bodies.
Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Fong KW, et al. J Cell Biol. 2013 Oct 14;203(1):149-64. doi: 10.1083/jcb.201303145. J Cell Biol. 2013. PMID: 24127217 Free PMC article.
References
- Leary D.J., Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. - PubMed
- Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. - PubMed
- Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nature Rev. Cancer. 2003;3:179–192. - PubMed