PRIDE: a public repository of protein and peptide identifications for the proteomics community - PubMed (original) (raw)
PRIDE: a public repository of protein and peptide identifications for the proteomics community
Philip Jones et al. Nucleic Acids Res. 2006.
Abstract
PRIDE, the 'PRoteomics IDEntifications database' (http://www.ebi.ac.uk/pride) is a database of protein and peptide identifications that have been described in the scientific literature. These identifications will typically be from specific species, tissues and sub-cellular locations, perhaps under specific disease conditions. Any post-translational modifications that have been identified on individual peptides can be described. These identifications may be annotated with supporting mass spectra. At the time of writing, PRIDE includes the full set of identifications as submitted by individual laboratories participating in the HUPO Plasma Proteome Project and a profile of the human platelet proteome submitted by the University of Ghent in Belgium. By late 2005 PRIDE is expected to contain the identifications and spectra generated by the HUPO Brain Proteome Project. Proteomics laboratories are encouraged to submit their identifications and spectra to PRIDE to support their manuscript submissions to proteomics journals. Data can be submitted in PRIDE XML format if identifications are included or mzData format if the submitter is depositing mass spectra without identifications. PRIDE is a web application, so submission, searching and data retrieval can all be performed using an internet browser. PRIDE can be searched by experiment accession number, protein accession number, literature reference and sample parameters including species, tissue, sub-cellular location and disease state. Data can be retrieved as machine-readable PRIDE or mzData XML (the latter for mass spectra without identifications), or as human-readable HTML.
Figures
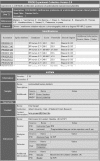
Figure 1
An example of PRIDE data in tabulated HTML format.
Figure 2
The PRIDE Advanced Search form.
Similar articles
- The PRIDE proteomics identifications database: data submission, query, and dataset comparison.
Jones P, Côté R. Jones P, et al. Methods Mol Biol. 2008;484:287-303. doi: 10.1007/978-1-59745-398-1_19. Methods Mol Biol. 2008. PMID: 18592187 - Using the PRIDE proteomics identifications database for knowledge discovery and data analysis.
Jones P, Martens L. Jones P, et al. Methods Mol Biol. 2010;604:297-307. doi: 10.1007/978-1-60761-444-9_20. Methods Mol Biol. 2010. PMID: 20013379 - PRIDE: the proteomics identifications database.
Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. Martens L, et al. Proteomics. 2005 Aug;5(13):3537-45. doi: 10.1002/pmic.200401303. Proteomics. 2005. PMID: 16041671 - Current trends in computational inference from mass spectrometry-based proteomics.
Webb-Robertson BJ, Cannon WR. Webb-Robertson BJ, et al. Brief Bioinform. 2007 Sep;8(5):304-17. doi: 10.1093/bib/bbm023. Epub 2007 Jun 20. Brief Bioinform. 2007. PMID: 17584764 Review. - Applications of diagonal chromatography for proteome-wide characterization of protein modifications and activity-based analyses.
Gevaert K, Impens F, Van Damme P, Ghesquière B, Hanoulle X, Vandekerckhove J. Gevaert K, et al. FEBS J. 2007 Dec;274(24):6277-89. doi: 10.1111/j.1742-4658.2007.06149.x. Epub 2007 Nov 16. FEBS J. 2007. PMID: 18021238 Review.
Cited by
- Development of data representation standards by the human proteome organization proteomics standards initiative.
Deutsch EW, Albar JP, Binz PA, Eisenacher M, Jones AR, Mayer G, Omenn GS, Orchard S, Vizcaíno JA, Hermjakob H. Deutsch EW, et al. J Am Med Inform Assoc. 2015 May;22(3):495-506. doi: 10.1093/jamia/ocv001. Epub 2015 Feb 28. J Am Med Inform Assoc. 2015. PMID: 25726569 Free PMC article. Review. - The mzIdentML data standard for mass spectrometry-based proteomics results.
Jones AR, Eisenacher M, Mayer G, Kohlbacher O, Siepen J, Hubbard SJ, Selley JN, Searle BC, Shofstahl J, Seymour SL, Julian R, Binz PA, Deutsch EW, Hermjakob H, Reisinger F, Griss J, Vizcaíno JA, Chambers M, Pizarro A, Creasy D. Jones AR, et al. Mol Cell Proteomics. 2012 Jul;11(7):M111.014381. doi: 10.1074/mcp.M111.014381. Epub 2012 Feb 27. Mol Cell Proteomics. 2012. PMID: 22375074 Free PMC article. - The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries.
Côté RG, Jones P, Apweiler R, Hermjakob H. Côté RG, et al. BMC Bioinformatics. 2006 Feb 28;7:97. doi: 10.1186/1471-2105-7-97. BMC Bioinformatics. 2006. PMID: 16507094 Free PMC article. - The use of neuroproteomics in drug abuse research.
Lull ME, Freeman WM, VanGuilder HD, Vrana KE. Lull ME, et al. Drug Alcohol Depend. 2010 Feb 1;107(1):11-22. doi: 10.1016/j.drugalcdep.2009.10.001. Epub 2009 Nov 17. Drug Alcohol Depend. 2010. PMID: 19926406 Free PMC article. Review. - NCBI Peptidome: a new repository for mass spectrometry proteomics data.
Ji L, Barrett T, Ayanbule O, Troup DB, Rudnev D, Muertter RN, Tomashevsky M, Soboleva A, Slotta DJ. Ji L, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D731-5. doi: 10.1093/nar/gkp1047. Epub 2009 Nov 26. Nucleic Acids Res. 2010. PMID: 19942688 Free PMC article.
References
- Orchard S., Hermjakob H., Apweiler R. The proteomics standards initiative. Proteomics. 2003;3:1374–1376. - PubMed
- Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. PRIDE: the proteomics identifications database. Proteomics. 2005;5:3537–3545. - PubMed
- Craig R., Cortens J.P., Beavis R.C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. - PubMed
- Deutsch E.W., Eng J.K., Zhang H., King N.L., Nesvizhskii A.I., Lin B., Lee H., Yi E.C., Ossola R., Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005;5:3497–3500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources