DBD: a transcription factor prediction database - PubMed (original) (raw)
DBD: a transcription factor prediction database
Sarah K Kummerfeld et al. Nucleic Acids Res. 2006.
Abstract
Regulation of gene expression influences almost all biological processes in an organism; sequence-specific DNA-binding transcription factors are critical to this control. For most genomes, the repertoire of transcription factors is only partially known. Hitherto transcription factor identification has been largely based on genome annotation pipelines that use pairwise sequence comparisons, which detect only those factors similar to known genes, or on functional classification schemes that amalgamate many types of proteins into the category of 'transcription factor'. Using a novel transcription factor identification method, the DBD transcription factor database fills this void, providing genome-wide transcription factor predictions for organisms from across the tree of life. The prediction method behind DBD identifies sequence-specific DNA-binding transcription factors through homology using profile hidden Markov models (HMMs) of domains. Thus, it is limited to factors that are homologus to those HMMs. The collection of HMMs is taken from two existing databases (Pfam and SUPERFAMILY), and is limited to models that exclusively detect transcription factors that specifically recognize DNA sequences. It does not include basal transcription factors or chromatin-associated proteins, for instance. Based on comparison with experimentally verified annotation, the prediction procedure is between 95% and 99% accurate. Between one quarter and one-half of our genome-wide predicted transcription factors represent previously uncharacterized proteins. The DBD (www.transcriptionfactor.org) consists of predicted transcription factor repertoires for 150 completely sequenced genomes, their domain assignments and the hand curated list of DNA-binding domain HMMs. Users can browse, search or download the predictions by genome, domain family or sequence identifier, view families of transcription factors based on domain architecture and receive predictions for a protein sequence.
Figures
Figure 1
Transcription factor prediction procedure. We begin with a set of proteins, shown as horizontal lines. For example, the initial set of proteins may be a whole proteome. Each sequence is searched against the SUPERFAMILY and Pfam HMM libraries. A domain is assigned to a particular protein when one of the HMMs matches a region of sequence with an _E_-value less than or equal to 0.001 for SUPERFAMILY or greater than or equal to the trusted cutoff for PFAM. Assigned domains are shown as coloured boxes where the colour indicates the family. For example, the small dark-blue boxes represent the Zinc finger C2H2 type DNA-binding domains. Proteins with at least one DNA-binding domain assigned are selected as putative transcription factors. The designation of DNA-binding is based on our manual curation of Pfam and SUPERFAMILY models.
Figure 2
DBD: Yeast predictions screen-shot. Each predicted transcription factor is listed with two rows for the SUPERFAMILY and PFAM domain architectures. Domains are represented as rectangles, coloured according to their family and horizontally located based on their position in the amino acid sequence. Clicking on a domain takes the user directly to that family in the relevant domain database. Proteins are ordered based on their domain architecture. For ease of navigation (in particular for large genomes), the list of transcription factors is split into pages with 50 entries per page by default. Users can navigate between pages using previous/next or clicking on a page number.
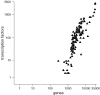
Figure 3
Number of genes in each of 151 genomes versus transcription factor predictions. The Number of genes (_x_-axis, log-scale) is plotted against the number of predicted transcription factors (_y_-axis, log-scale). Each splice variant is counted independently. (See database website for a list of genomes considered.)
Similar articles
- DBD--taxonomically broad transcription factor predictions: new content and functionality.
Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. Wilson D, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D88-92. doi: 10.1093/nar/gkm964. Epub 2007 Dec 11. Nucleic Acids Res. 2008. PMID: 18073188 Free PMC article. - webPRC: the Profile Comparer for alignment-based searching of public domain databases.
Brandt BW, Heringa J. Brandt BW, et al. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W48-52. doi: 10.1093/nar/gkp279. Epub 2009 May 6. Nucleic Acids Res. 2009. PMID: 19420063 Free PMC article. - Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure.
Gough J, Karplus K, Hughey R, Chothia C. Gough J, et al. J Mol Biol. 2001 Nov 2;313(4):903-19. doi: 10.1006/jmbi.2001.5080. J Mol Biol. 2001. PMID: 11697912 - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - Genomic repertoires of DNA-binding transcription factors across the tree of life.
Charoensawan V, Wilson D, Teichmann SA. Charoensawan V, et al. Nucleic Acids Res. 2010 Nov;38(21):7364-77. doi: 10.1093/nar/gkq617. Epub 2010 Jul 30. Nucleic Acids Res. 2010. PMID: 20675356 Free PMC article. Review.
Cited by
- Comparative Analysis of Prostate Cancer Gene Regulatory Networks via Hub Type Variation.
Khosravi P, Gazestani VH, Akbarzadeh M, Mirkhalaf S, Sadeghi M, Goliaei B. Khosravi P, et al. Avicenna J Med Biotechnol. 2015 Jan-Mar;7(1):8-15. Avicenna J Med Biotechnol. 2015. PMID: 25926947 Free PMC article. - In silico method for modelling metabolism and gene product expression at genome scale.
Lerman JA, Hyduke DR, Latif H, Portnoy VA, Lewis NE, Orth JD, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Zengler K, Palsson BO. Lerman JA, et al. Nat Commun. 2012 Jul 3;3:929. doi: 10.1038/ncomms1928. Nat Commun. 2012. PMID: 22760628 Free PMC article. - Identification of lung-specific genes by meta-analysis of multiple tissue RNA-seq data.
Xiong M, Heruth DP, Zhang LQ, Ye SQ. Xiong M, et al. FEBS Open Bio. 2016 Jun 16;6(7):774-81. doi: 10.1002/2211-5463.12089. eCollection 2016 Jul. FEBS Open Bio. 2016. PMID: 27398317 Free PMC article. - Universal patterns of purifying selection at noncoding positions in bacteria.
Molina N, van Nimwegen E. Molina N, et al. Genome Res. 2008 Jan;18(1):148-60. doi: 10.1101/gr.6759507. Epub 2007 Nov 21. Genome Res. 2008. PMID: 18032729 Free PMC article. - Molecular Analysis of UV-C Induced Resveratrol Accumulation in Polygonum cuspidatum Leaves.
Liu Z, Xu J, Wu X, Wang Y, Lin Y, Wu D, Zhang H, Qin J. Liu Z, et al. Int J Mol Sci. 2019 Dec 7;20(24):6185. doi: 10.3390/ijms20246185. Int J Mol Sci. 2019. PMID: 31817915 Free PMC article.
References
- Salgado H., Gama-Castro S., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., Collado-Vides J. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 2004;32:D303–D306. - PMC - PubMed
- Kanamori M., Konno H., Osato N., Kawai J., Hayashizaki Y., Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem. Biophys. Res. Commun. 2004;322:787–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources