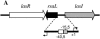The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter - PubMed (original) (raw)
The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter
Giordano Rampioni et al. J Bacteriol. 2006 Jan.
Abstract
A mutation in the rsaL gene of Pseudomonas aeruginosa produces dramatically higher amounts of N-acyl homoserine lactone with respect to the wild type, highlighting the key role of this negative regulator in controlling quorum sensing (QS) in this opportunistic pathogen. The DNA binding site of the RsaL protein on the rsaL-lasI bidirectional promoter partially overlaps the binding site of the LasR protein, consistent with the hypothesis that RsaL and LasR could be in binding competition on this promoter. This is the first direct demonstration that RsaL acts as a QS negative regulator by binding to the lasI promoter.
Figures
FIG. 1.
Genetic organization of the P. aeruginosa las quorum sensing locus and detail of the _rsaL_-lasI bidirectional promoter (below). The lasR, rsaL, and lasI genes are depicted as arrows. Bent arrows indicate the _rsaL_- and _lasI_- divergent transcripts. The open box and the gray box indicate the LasR and RsaL binding sites, respectively. Numbers indicate the center of the LasR and RsaL binding sites with respect to the lasI transcription starting site (22).
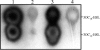
FIG. 2.
TLC analysis of acyl-HSL produced by parent strain P. aeruginosa PAO1 and its _rsaL_-negative mutant derivative visualized by overlaying with acyl-HSL sensor strain E. coli(pSB1075) (30). Lane 1, synthetic 3OC12-HSL and 3OC10-HSL (0.8 μg); lane 2, PAO1; lane 3, PAO1 rsaL; lane 4, PAO1 rsaL carrying plasmid pPSRsaLPAO. Acyl-HSLs were extracted from spent supernatants, and a volume corresponding to 8 × 108 CFU was loaded for the TLC assay.
FIG. 3.
Results of overexpression and purification of recombinant RsaLPAO6H are shown. (A) Analysis of protein samples from key steps of purification by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane 1, PageRuler protein ladder (Fermentas Inc.) (only molecular mass markers from 50 kDa to 10 kDa are indicated); lane 2, uninduced whole-cell extract; lane 3, induced whole-cell extract; lane 4, insoluble cell extract; lane 5, soluble cell extract; lane 6, pooled fractions eluted at 250 mM imidazole. The arrow indicates the purified RsaLPAO6H protein (10.8 kDa). (B) Western blot with anti-six-histidine antibodies of a sodium dodecyl sulfide-polyacrylamide gel identical to that shown in panel A.
FIG. 4.
EMSA of RsaLPAO6H binding to the lasI promoter. The 32P-labeled DNA fragment contained nucleotides −83 to +20 with respect to the lasI transcriptional starting site. RsaLPAO6H concentrations (nM) are indicated below the lanes. The probe concentration for each sample was 2 nM. As specific and unspecific competitors, unlabeled probe (100 nM) and calf thymus DNA (1 μg) were added to lane 9 and 10, respectively. The arrow indicates the unshifted DNA probe.
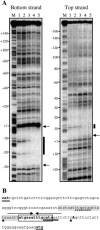
FIG. 5.
(A) DNase I footprints of RsaLPAO6H on the _rsaL_-lasI intergenic region. The plasmids p_PlasI_5′ (for labeling of the bottom strand) and p_PlasI_3′ (for labeling of the top strand) were utilized to generate EcoRI/SacI fragments used as probes (Tables 1 and 2). The probes were mixed with different amounts of RsaLPAO6H protein prior to DNase I digestion. Thick lines indicate the regions showing specific protection by RsaLPAO6H; arrows indicate hypersensitive sites. All numbering is in reference to the transcriptional starting site from the lasI promoter (23). M, Maxam and Gilbert sequencing reactions (A+G); lane 1, no RsaLPAO6H added; lanes 2 to 5, RsaLPAO6H added to a final concentration of 0.05, 0.5, 5, or 50 μM, respectively. (B) Sequence of the _rsaL_-lasI intergenic region. The lasI ATG starting codon is boldface and underlined, and the nucleotides complementary to the starting codon of rsaL (CAT) are boldface and double underlined. The lasI transcriptional starting site is boldface and capitalized (23). The sequence protected by RsaL in the DNase I protection assay is boldface and boxed, and hypersensitive sites are indicated by triangles. The sequence protected by LasR is gray shaded. The 5′-AAnTTATGnAA-3′ inverted repeats are indicated by arrows. The potential σ70-dependent −35 and −10 consensus sequences are indicated by dashed and solid thick lines, respectively.
Similar articles
- RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa.
Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. Rampioni G, et al. Mol Microbiol. 2007 Dec;66(6):1557-65. doi: 10.1111/j.1365-2958.2007.06029.x. Mol Microbiol. 2007. PMID: 18045385 - The multi-output incoherent feedforward loop constituted by the transcriptional regulators LasR and RsaL confers robustness to a subset of quorum sensing genes in Pseudomonas aeruginosa.
Bondí R, Longo F, Messina M, D'Angelo F, Visca P, Leoni L, Rampioni G. Bondí R, et al. Mol Biosyst. 2017 Jun 1;13(6):1080-1089. doi: 10.1039/c7mb00040e. Epub 2017 May 3. Mol Biosyst. 2017. PMID: 28466892 - Functional characterization of the quorum sensing regulator RsaL in the plant-beneficial strain Pseudomonas putida WCS358.
Rampioni G, Bertani I, Pillai CR, Venturi V, Zennaro E, Leoni L. Rampioni G, et al. Appl Environ Microbiol. 2012 Feb;78(3):726-34. doi: 10.1128/AEM.06442-11. Epub 2011 Nov 23. Appl Environ Microbiol. 2012. PMID: 22113916 Free PMC article. - Impact of quorum sensing on fitness of Pseudomonas aeruginosa.
Heurlier K, Dénervaud V, Haas D. Heurlier K, et al. Int J Med Microbiol. 2006 Apr;296(2-3):93-102. doi: 10.1016/j.ijmm.2006.01.043. Epub 2006 Feb 28. Int J Med Microbiol. 2006. PMID: 16503417 Review. - The virtue of temperance: built-in negative regulators of quorum sensing in Pseudomonas.
Venturi V, Rampioni G, Pongor S, Leoni L. Venturi V, et al. Mol Microbiol. 2011 Dec;82(5):1060-70. doi: 10.1111/j.1365-2958.2011.07890.x. Epub 2011 Nov 7. Mol Microbiol. 2011. PMID: 22060261 Review.
Cited by
- Negative regulation of bacterial quorum sensing tunes public goods cooperation.
Gupta R, Schuster M. Gupta R, et al. ISME J. 2013 Nov;7(11):2159-68. doi: 10.1038/ismej.2013.109. Epub 2013 Jul 4. ISME J. 2013. PMID: 23823496 Free PMC article. - The multiple signaling systems regulating virulence in Pseudomonas aeruginosa.
Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. Jimenez PN, et al. Microbiol Mol Biol Rev. 2012 Mar;76(1):46-65. doi: 10.1128/MMBR.05007-11. Microbiol Mol Biol Rev. 2012. PMID: 22390972 Free PMC article. Review. - The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor.
Rajamani S, Bauer WD, Robinson JB, Farrow JM 3rd, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. Rajamani S, et al. Mol Plant Microbe Interact. 2008 Sep;21(9):1184-92. doi: 10.1094/MPMI-21-9-1184. Mol Plant Microbe Interact. 2008. PMID: 18700823 Free PMC article. - Negative Regulation of Violacein Biosynthesis in Chromobacterium violaceum.
Devescovi G, Kojic M, Covaceuszach S, Cámara M, Williams P, Bertani I, Subramoni S, Venturi V. Devescovi G, et al. Front Microbiol. 2017 Mar 7;8:349. doi: 10.3389/fmicb.2017.00349. eCollection 2017. Front Microbiol. 2017. PMID: 28326068 Free PMC article. - Social cheating in Pseudomonas aeruginosa quorum sensing.
Sandoz KM, Mitzimberg SM, Schuster M. Sandoz KM, et al. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15876-81. doi: 10.1073/pnas.0705653104. Epub 2007 Sep 26. Proc Natl Acad Sci U S A. 2007. PMID: 17898171 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases