The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases - PubMed (original) (raw)
Comparative Study
. 2006 Jan 10;45(1):121-30.
doi: 10.1021/bi051787e.
Affiliations
- PMID: 16388587
- PMCID: PMC2570371
- DOI: 10.1021/bi051787e
Comparative Study
The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases
Craig W Vander Kooi et al. Biochemistry. 2006.
Abstract
Prp19 is an essential splicing factor and a member of the U-box family of E3 ubiquitin ligases. Prp19 forms a tetramer via a central coiled-coil domain. Here, we show the U-box domain of Prp19 exists as a dimer within the context of the Prp19 tetramer. A high-resolution structure of the homodimeric state of the Prp19 U-box was determined by X-ray crystallography. Mutation of the U-box dimer interface abrogates U-box dimer formation and is lethal in vivo. The structure of the U-box dimer enables construction of a complete model of Prp19 providing insights into how the tetrameric protein functions as an E3 ligase. Finally, comparison of the Prp19 U-box homodimer with the heterodimeric complex of BRCA1/BARD1 RING-finger domains uncovers a common architecture for a family of oligomeric U-box and RING-finger E3 ubiquitin ligases, which has mechanistic implications for E3 ligase-mediated polyubiquitination and E4 polyubiquitin ligases.
Figures
Figure 1
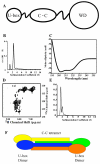
The U-box domain of Prp19 exists as a functional dimer. A) Prp19 contains three domains: an N-terminal U-box, a central tetrameric coiled-coil, and a C-terminal WD-40 repeat domain. B) Analytical ultracetrifugation reveals that the coiled-coil domain drives the formation of a single functional tetramer in the context of a construct containing both U-box and coiled-coil domains. C) The construct containing U-box and coiled-coil domains is well structured with primarily helical content. D) NMR reveals that the U-box domain is integrally connected to Prp19 and not flexibly linked, as is the case for the C-terminal WD40 repeats. E) The U-box of Prp19 exists in a monomer/dimer equilibrium. F) A model of the U-box and coiled-coil regions of Prp19 showing the symmetric U-box dimers formed around the coiled-coil tetramer.
Figure 2
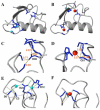
Basis for Prp19 dimerization A) Ribbon diagram of the Prp19 U-box dimer. Molecular graphics were generated using MOLMOL (48). B) Prp19 contains a central hydrophobic patch at the dimer interface composed of four long-chain hydrophobic residues, shown with a zoom showing the electron density at the dimer interface. C) Representative 2FO-FC electron density map of the dimer interface contoured at 2σ (generated with PYMOL;
). D) Mutation of the central hydrophobic L15 to glutamate disrupts the ability of the U-box to dimerize. AU profile of Prp19(1-73) L15E shows only one peak with a sedimentation coeffiecient consistent with the monomeric species. E) 15N-1H HSQC of the same construct shows excellent line width and chemical shift dispersion, characteristics of a well folded domain. Additionally, the chemical shifts are virtually superimposable with the wild-type protein (19) demonstrating that the L15E mutation causes minimal structural perturbation and adopts a native-like structure. F) Mutations to the residues in the dimer interface abrogate Prp19 function in vivo either in rescuing a temperature sensitive strain (Upper panel) or rescuing growth for the null allele (Lower panel).
Figure 3
Stabilization centers of the U-box domain. A) U-box domain is stabilized by two networks of hydrogen bonds and salt bridges in Prp19. B) In contrast the RING-finger domain of Rag1 (1RMD) is stabilized by chelation of two zinc atoms. C, D) The C-terminal stabilization center of Prp19 is much more compact and largely recapitulates the organization of the RING-finger center (D), but with Asp38 extended to occupy the position of the central zinc atom. E) The N-terminal stabilization center of the U-box contains an extended network of inter-residue interactions including many between side chain and main chain amides. Distances between potential hydrogen bond donors and acceptors are color coded: d < 3.0Å as green, 3.0< d <3.25 as pink, 3.25< d <3.50 as tan. F) The zinc mediated RING-finger contains the four zinc-sulfur bonds.
Figure 4
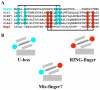
Comparison of related E3 ligase domain stabilization A) Multiple sequence alignment of Prp19, Rag1, and three human Miz-finger domains. Residues contributing to the N-terminal center of the U-box are highlighted in blue. Residues contributing to the C-terminal center of the RING-finger domain are highlighted in red. B) Schematic of the cross-braced stabilization of U-box and RING-finger proteins with hydrogen-bonding and zinc centers in cyan and red, respectively. Both domains contain a core helix (large rectangle) and three short β-strands (small rectangles). The cross-brace is mediated through the central strand, represented by the dotted line connecting the two stabilization centers. The Miz-finger domain found in related SUMO E3 ligases appears to represent a hybrid of these domains.
Figure 5
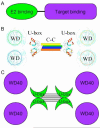
Model of the quaternary structure of Prp19. A) Prp19 exists as a functional tetramer with dimeric U-box domains and flexibly attached WD40 domains. WD40 repeat domain modeled from Groucho/Tle1 (49). B) The basic domain structure of an E3 ligase requires E2 and substrate recruitment domains. C) The quaternary structure of Prp19 suggests a common architecture for oligomeric E3 ubiquitin ligases that brings the E2 recruitment domain into close proximity with a substrate recognition domain.
Figure 6
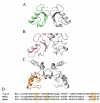
The Prp19 dimer as a general model for oligomeric E3 ligases. Comparison of the dimerization interfaces of A) Prp19 B) BRCA1/BARD1 (1JM7) and C) Rag1 (1RMD), respectively, show different characterized dimerization interfaces. The U-box or RING-finger dimerization interface appears structurally conserved between Prp19 and BRCA1 D) Multiple sequence alignment of dimeric RING-finger domains of Mdm2 and MdmX with Prp19. The four hydrophobic residues at the Prp19 dimer interface are highlighted.
Similar articles
- Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Zhang M, et al. Mol Cell. 2005 Nov 23;20(4):525-38. doi: 10.1016/j.molcel.2005.09.023. Mol Cell. 2005. PMID: 16307917 - Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein.
Xu Z, Devlin KI, Ford MG, Nix JC, Qin J, Misra S. Xu Z, et al. Biochemistry. 2006 Apr 18;45(15):4749-59. doi: 10.1021/bi0601508. Biochemistry. 2006. PMID: 16605243 - Symmetry and asymmetry of the RING-RING dimer of Rad18.
Huang A, Hibbert RG, de Jong RN, Das D, Sixma TK, Boelens R. Huang A, et al. J Mol Biol. 2011 Jul 15;410(3):424-35. doi: 10.1016/j.jmb.2011.04.051. Epub 2011 Apr 27. J Mol Biol. 2011. PMID: 21549715 - RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination.
Metzger MB, Pruneda JN, Klevit RE, Weissman AM. Metzger MB, et al. Biochim Biophys Acta. 2014 Jan;1843(1):47-60. doi: 10.1016/j.bbamcr.2013.05.026. Epub 2013 Jun 6. Biochim Biophys Acta. 2014. PMID: 23747565 Free PMC article. Review. - Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination.
Christensen DE, Klevit RE. Christensen DE, et al. FEBS J. 2009 Oct;276(19):5381-9. doi: 10.1111/j.1742-4658.2009.07249.x. Epub 2009 Aug 27. FEBS J. 2009. PMID: 19712108 Free PMC article. Review.
Cited by
- The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage.
Lu X, Legerski RJ. Lu X, et al. Biochem Biophys Res Commun. 2007 Mar 23;354(4):968-74. doi: 10.1016/j.bbrc.2007.01.097. Epub 2007 Jan 26. Biochem Biophys Res Commun. 2007. PMID: 17276391 Free PMC article. - UBC13 is an RNF213-associated E2 ubiquitin-conjugating enzyme, and Lysine 63-linked ubiquitination by the RNF213-UBC13 axis is responsible for angiogenic activity.
Habu T, Harada KH. Habu T, et al. FASEB Bioadv. 2021 Jan 20;3(4):243-258. doi: 10.1096/fba.2019-00092. eCollection 2021 Apr. FASEB Bioadv. 2021. PMID: 33842849 Free PMC article. - Structural basis for conformational equilibrium of the catalytic spliceosome.
Wilkinson ME, Fica SM, Galej WP, Nagai K. Wilkinson ME, et al. Mol Cell. 2021 Apr 1;81(7):1439-1452.e9. doi: 10.1016/j.molcel.2021.02.021. Epub 2021 Mar 10. Mol Cell. 2021. PMID: 33705709 Free PMC article. - Intra molecular interactions in the regulation of p53 pathway.
Chen J. Chen J. Transl Cancer Res. 2016 Dec;5(6):639-649. doi: 10.21037/tcr.2016.09.23. Transl Cancer Res. 2016. PMID: 30613472 Free PMC article. - In vitro characterization of six STUB1 variants in spinocerebellar ataxia 16 reveals altered structural properties for the encoded CHIP proteins.
Pakdaman Y, Sanchez-Guixé M, Kleppe R, Erdal S, Bustad HJ, Bjørkhaug L, Haugarvoll K, Tzoulis C, Heimdal K, Knappskog PM, Johansson S, Aukrust I. Pakdaman Y, et al. Biosci Rep. 2017 Apr 28;37(2):BSR20170251. doi: 10.1042/BSR20170251. Print 2017 Apr 30. Biosci Rep. 2017. PMID: 28396517 Free PMC article.
References
- Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–8. - PubMed
- Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23:1958–71. - PubMed
- Hochstrasser M. Ubiquitin and intracellular protein degradation. Curr Opin Cell Biol. 1992;4:1024–31. - PubMed
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM55420/GM/NIGMS NIH HHS/United States
- P30 ES000267/ES/NIEHS NIH HHS/United States
- R01GM75156/GM/NIGMS NIH HHS/United States
- T32 GM008320/GM/NIGMS NIH HHS/United States
- R01 GM055420-12/GM/NIGMS NIH HHS/United States
- T32 CA009385/CA/NCI NIH HHS/United States
- R01 GM062112-04/GM/NIGMS NIH HHS/United States
- R01 GM062112-03/GM/NIGMS NIH HHS/United States
- R01GM62112/GM/NIGMS NIH HHS/United States
- T32 GM008320-13/GM/NIGMS NIH HHS/United States
- P30 ES000267-37/ES/NIEHS NIH HHS/United States
- R01 GM055420-11/GM/NIGMS NIH HHS/United States
- T32 GM008320-12/GM/NIGMS NIH HHS/United States
- T32 CA009385-21/CA/NCI NIH HHS/United States
- P30 ES000267-38/ES/NIEHS NIH HHS/United States
- T32CA09385/CA/NCI NIH HHS/United States
- R01 GM062112/GM/NIGMS NIH HHS/United States
- R01 GM075156-01/GM/NIGMS NIH HHS/United States
- T32GM08320/GM/NIGMS NIH HHS/United States
- R01 GM075156/GM/NIGMS NIH HHS/United States
- R01 GM055420/GM/NIGMS NIH HHS/United States
- R01 GM062112-05/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous