Identification of eukaryotic open reading frames in metagenomic cDNA libraries made from environmental samples - PubMed (original) (raw)
Identification of eukaryotic open reading frames in metagenomic cDNA libraries made from environmental samples
Susan Grant et al. Appl Environ Microbiol. 2006 Jan.
Abstract
Here we describe the application of metagenomic technologies to construct cDNA libraries from RNA isolated from environmental samples. RNAlater (Ambion) was shown to stabilize RNA in environmental samples for periods of at least 3 months at -20 degrees C. Protocols for library construction were established on total RNA extracted from Acanthamoeba polyphaga trophozoites. The methodology was then used on algal mats from geothermal hot springs in Tengchong county, Yunnan Province, People's Republic of China, and activated sludge from a sewage treatment plant in Leicestershire, United Kingdom. The Tenchong libraries were dominated by RNA from prokaryotes, reflecting the mainly prokaryote microbial composition. The majority of these clones resulted from rRNA; only a few appeared to be derived from mRNA. In contrast, many clones from the activated sludge library had significant similarity to eukaryote mRNA-encoded protein sequences. A library was also made using polyadenylated RNA isolated from total RNA from activated sludge; many more clones in this library were related to eukaryotic mRNA sequences and proteins. Open reading frames (ORFs) up to 378 amino acids in size could be identified. Some resembled known proteins over their full length, e.g., 36% match to cystatin, 49% match to ribosomal protein L32, 63% match to ribosomal protein S16, 70% to CPC2 protein. The methodology described here permits the polyadenylated transcriptome to be isolated from environmental samples with no knowledge of the identity of the microorganisms in the sample or the necessity to culture them. It has many uses, including the identification of novel eukaryotic ORFs encoding proteins and enzymes.
Figures
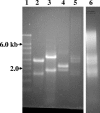
FIG. 1.
Typical results of total RNA extraction from named species and environmental samples electrophoresed on a 1.2% denaturing formaldehyde agarose gel. Lane 1, RNA size markers (Ambion Millennium markers); lane 2, Escherichia coli; lane 3, Saccharomyces cerevisiae; lane 4, Acanthamoeba polyphaga; lanes 5 and 6, activated sludge.
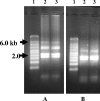
FIG. 2.
Effect of storage in RNAlater on the stability of total RNA extracted from Acanthamoeba polyphaga. (A) TBE 1.2% agarose gel. Lane 1, RNA size markers (Millennium markers; Ambion); lanes 2 and 3, total RNA from a fresh culture of Acanthamoeba polyphaga. (B) Same as panel A, but lane 2, total RNA from A. polyphaga stored in RNAlater for 10 days at ambient temperature; lane 3, total RNA from A. polyphaga stored for 10 days at 4°C.
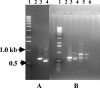
FIG. 3.
Results of reverse transcription and LD-PCR on RNA from different sources to produce cDNAs run out on TBE 1.2% agarose gels. (A) Lane 1, 1-kb DNA ladder (Invitrogen); lane 2, negative control; lane 3, product from the human placental mRNA provided as a control in the Smart cDNA Library construction kit. (B) Lane 1, 1-kb ladder; lanes 2 to 5, product from total RNA extracted from Acanthamoeba polyphaga. (C) Same as panel B, showing product from the LP4 Chinese algal mat. (D) Same as panel B, showing product from activated sludge total RNA.
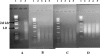
FIG. 4.
18S rRNA gene amplification from algal mat samples and activated sludge. (A) Lane 1, 1-kb ladder; lane 2, negative control; lane 3, Acanthamoeba polyphaga positive control; lane 4, product from activated sludge. (B) Lane 1, 1-kb ladder; lane 2, negative control; lane 3, Acanthamoeba polyphaga positive control; lane 4, algal mat LP4; lanes 5 and 6, product from two different DNA extracts from algal mat TC2.
Similar articles
- Constructing high complexity synthetic libraries of long ORFs using in vitro selection.
Cho G, Keefe AD, Liu R, Wilson DS, Szostak JW. Cho G, et al. J Mol Biol. 2000 Mar 24;297(2):309-19. doi: 10.1006/jmbi.2000.3571. J Mol Biol. 2000. PMID: 10715203 - Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches.
Massana R, Balagué V, Guillou L, Pedrós-Alió C. Massana R, et al. FEMS Microbiol Ecol. 2004 Nov 1;50(3):231-43. doi: 10.1016/j.femsec.2004.07.001. FEMS Microbiol Ecol. 2004. PMID: 19712363 - A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples.
Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. Stoeck T, et al. Protist. 2006 Feb;157(1):31-43. doi: 10.1016/j.protis.2005.10.004. Epub 2006 Jan 23. Protist. 2006. PMID: 16431157 - [Organization and evolution of the yeast Saccharomyces cerevisiae genome and the problem of orphan open reading frames].
Mackiewicz P. Mackiewicz P. Postepy Biochem. 2000;46(4):291-8. Postepy Biochem. 2000. PMID: 11449963 Review. Polish. No abstract available. - Discovering Protein-Coding Genes from the Environment: Time for the Eukaryotes?
Marmeisse R, Kellner H, Fraissinet-Tachet L, Luis P. Marmeisse R, et al. Trends Biotechnol. 2017 Sep;35(9):824-835. doi: 10.1016/j.tibtech.2017.02.003. Epub 2017 Mar 7. Trends Biotechnol. 2017. PMID: 28279485 Review.
Cited by
- A Novel 1,3-Β-Glucanase Gene from the Metagenomic Expression Library of Achatina Fulica's Digestive Gland.
Kurniawati M, Purkan, Sumarsih S, Baktir A. Kurniawati M, et al. Iran J Pharm Res. 2020 Summer;19(3):483-493. doi: 10.22037/ijpr.2020.1101172. Iran J Pharm Res. 2020. PMID: 33680046 Free PMC article. - Structural and functional diversity of free-living microorganisms in reef surface, Kra island, Thailand.
Somboonna N, Wilantho A, Assawamakin A, Monanunsap S, Sangsrakru D, Tangphatsornruang S, Tongsima S. Somboonna N, et al. BMC Genomics. 2014 Jul 18;15:607. doi: 10.1186/1471-2164-15-607. BMC Genomics. 2014. PMID: 25037613 Free PMC article. - Fungal community analysis by high-throughput sequencing of amplified markers--a user's guide.
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. Lindahl BD, et al. New Phytol. 2013 Jul;199(1):288-299. doi: 10.1111/nph.12243. Epub 2013 Mar 28. New Phytol. 2013. PMID: 23534863 Free PMC article. Review. - Gene transcription in Lactarius quietus-Quercus petraea ectomycorrhizas from a forest soil.
Courty PE, Poletto M, Duchaussoy F, Buée M, Garbaye J, Martin F. Courty PE, et al. Appl Environ Microbiol. 2008 Nov;74(21):6598-605. doi: 10.1128/AEM.00584-08. Epub 2008 Sep 12. Appl Environ Microbiol. 2008. PMID: 18791033 Free PMC article. - Metagenomics and metatranscriptomics as potential driving forces for the exploration of diversity and functions of micro-eukaryotes in soil.
Yadav BNS, Sharma P, Maurya S, Yadav RK. Yadav BNS, et al. 3 Biotech. 2023 Dec;13(12):423. doi: 10.1007/s13205-023-03841-3. Epub 2023 Nov 30. 3 Biotech. 2023. PMID: 38047037 Free PMC article. Review.
References
- Abramczyk, D., M. Tchorzewski, and N. Grankowski. 2003. Non-AUG translation initiation of mRNA encoding acidic ribosomal P2A protein in Candida albicans. Yeast 20:1045-1052. - PubMed
- Amaral Zettler, L. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases