A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis - PubMed (original) (raw)
. 2006 Jan 10;103(2):359-64.
doi: 10.1073/pnas.0508883103. Epub 2006 Jan 3.
Lene Heegaard Madsen, Simona Radutoiu, Mirela Frantescu, Esben M H Quistgaard, Hiroki Miwa, J Allan Downie, Euan K James, Hubert H Felle, Line Lindegaard Haaning, Torben Heick Jensen, Shusei Sato, Yasukazu Nakamura, Satoshi Tabata, Niels Sandal, Jens Stougaard
Affiliations
- PMID: 16407163
- PMCID: PMC1326171
- DOI: 10.1073/pnas.0508883103
A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis
Norihito Kanamori et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Nuclear-cytoplasmic partitioning and traffic between cytoplasmic and nuclear compartments are fundamental processes in eukaryotic cells. Nuclear pore complexes mediate transport of proteins, RNAs and ribonucleoprotein particles in and out of the nucleus. Here we present positional cloning of a plant nucleoporin gene, Nup133, essential for a symbiotic signal transduction pathway shared by Rhizobium bacteria and mycorrhizal fungi. Mutation of Nup133 results in a temperature sensitive nodulation deficient phenotype and absence of mycorrhizal colonization. Root nodules developing with reduced frequency at permissive temperatures are ineffective and electron microscopy show that Rhizobium bacteria are not released from infection threads. Measurement of ion fluxes using a calcium-sensitive dye show that Nup133 is required for the Ca2+ spiking normally detectable within minutes after application of purified rhizobial Nod-factor signal molecules to root hairs. Localization of NUP133 in the nuclear envelope of root cells and root hair cells shown with enhanced yellow fluorescent protein fusion proteins suggests a novel role for NUP133 nucleoporins in a rapid nuclear-cytoplasmic communication after host-plant recognition of symbiotic microbes. Our results identify a component of an intriguing signal process requiring interaction at the cell plasma membrane and at intracellular nuclear and plastid organelle-membranes to induce a second messenger.
Figures
Fig. 1.
Phenotype of nup133 mutants and genetic complementation. (A) Wild-type root nodules. (B) Ineffective nodule on _nup133_-1 inoculated with the M. loti TONO strain. (C) Root hair curling on a wild-type seedling inoculated with M. loti. (D) Root hair swelling on _nup133_-1 inoculated with M. loti. (E) Nodules on transgenic root showing complementation of _nup133_-2. (F) Section of mature wild-type root nodule. (G) Section of _nup133_-2 nodule. (H_–_J) Electron micrographs of an ineffective nodule showing infection threads (arrow) and M. loti bacteria within enlarged infection droplet structures (arrow). (K and L) Electron micrographs of a wild-type nodule showing a branched infection thread releasing an M. loti containing infection droplet (white arrow) and bacteroids (black arrow) surrounded by peribacteroid membrane (arrowhead). The symbiosome consists of bacteroids enclosed within a peribacteroid membrane. (Scale bars: 1 mm in A, B, and G and 50 μm in C_–_F and H.)
Fig. 2.
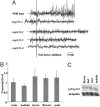
Calcium oscillations in root hairs in response to purified Nod-factor. (A) Each trace is from a single root hair using seedlings of wild-type L. japonicus or mutants carrying the _nup133_-1, _nup133_-2, _nup133_-3 or _nup133_-4 alleles. Seven, four, six, and three root hairs were analyzed for each allele, respectively. An additional 10 analyses were done with nup133-3 grown at 10°C and assayed at 15°C, but the traces were similar to that shown. Root hairs were injected with the calcium-sensitive dye Oregon green-488 BAPTA-1 and, after ≈20 min, Nod factor was added at 10-8 M. The data are graphed showing typical traces of the first derivatives of fluorescence intensity (in arbitrary units) between 5 s sequential time points. There was no observed calcium spiking in any of the mutants under the conditions tested. (B and C) Transcript level of Nup133 in wild-type plants. (B) Quantitative RT-PCR results are shown as fold increase compared to roots. Error bars represent a 95% confidence interval. (C) Northern hybridization showing similar expression levels of Nup133 in different Lotus tissues
Fig. 3.
Alignment of the N-terminal 50–535 residues of Lotus NUP133 to human Nup133 (top sequence). Amino acid residues marked in gray are identical. Propeller blades are boxed and β strands named from 1 (A–D) to 7 (A–D) after position in the human Nup133 propeller are shown below. Human Nup133 strand annotations are depicted above the Lotus NUP133 strand predictions. Although Lotus NUP133 has quite a few insertions compared to human Nup133, the N-terminal domain of Lotus NUP133 is predicted to adopt a seven-bladed β-propeller fold, similar to the human Nup133 propeller.
Fig. 4.
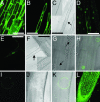
Subcellular localization of eYFP–NUP133 fusion protein in the nuclear rim. (A and B) Fluorescent image of a control eYFP localization in _nup133_-1.(C) Light microscopy of B with nucleus marked by arrow and highlighted in insert. (D, E, H, I, K, and L) Fluorescent images of eYFP–NUP133 fusion protein showing nuclear rim localization in _nup133_-1 mutant background. (E_–_K) Single cell localisation of eYFP–NUP133 in the nuclear rim. (E) Root hair cell showing nuclear rim localization of eYFP–NUP133. (F) Corresponding light microscopy with nucleus marked by arrow and highlighted in Inset. (H) Fluorescent overlay of G light microscopy with nuclei marked by arrows. Note the surface view of one nucleus showing punctate localization of eYFP–NUP133 fusion protein. (K) Overlay of corresponding fluorescent image of eYFP–NUP133 (I) and light microscopy image (J). Note the punctate eYFP–NUP133 localization. (L) Localization of eYFP–NUP133 in nuclear rims of cells in a transgenic root tip; note the cell files. (A and D) Epidermal cells. (B, C, E, and F) Root hair cells. (Scale bar: 50 μm.)
Similar articles
- NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus.
Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, Kouchi H, Murooka Y, Szczyglowski K, Downie JA, Parniske M, Hayashi M, Kawaguchi M. Saito K, et al. Plant Cell. 2007 Feb;19(2):610-24. doi: 10.1105/tpc.106.046938. Epub 2007 Feb 16. Plant Cell. 2007. PMID: 17307929 Free PMC article. - CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis.
Yano K, Shibata S, Chen WL, Sato S, Kaneko T, Jurkiewicz A, Sandal N, Banba M, Imaizumi-Anraku H, Kojima T, Ohtomo R, Szczyglowski K, Stougaard J, Tabata S, Hayashi M, Kouchi H, Umehara Y. Yano K, et al. Plant J. 2009 Oct;60(1):168-80. doi: 10.1111/j.1365-313X.2009.03943.x. Epub 2009 Jun 5. Plant J. 2009. PMID: 19508425 - Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program.
Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolín-Llovera M, Parniske M, Oldroyd GE, Downie AJ, Karas B, Szczyglowski K. Kosuta S, et al. Plant J. 2011 Sep;67(5):929-40. doi: 10.1111/j.1365-313X.2011.04645.x. Epub 2011 Jul 1. Plant J. 2011. PMID: 21595760 - [Calcium oscillations and Nod signal transduction in Rhizobium-legume symbiosis].
Gough C. Gough C. J Soc Biol. 2001;195(3):297-302. doi: 10.1051/jbio/2001195030297. J Soc Biol. 2001. PMID: 11833467 Review. French. - Root Development and Endosymbioses: DELLAs Lead the Orchestra.
Fonouni-Farde C, Diet A, Frugier F. Fonouni-Farde C, et al. Trends Plant Sci. 2016 Nov;21(11):898-900. doi: 10.1016/j.tplants.2016.08.012. Epub 2016 Sep 22. Trends Plant Sci. 2016. PMID: 27666515 Review.
Cited by
- The Arabidopsis nuclear pore and nuclear envelope.
Meier I, Brkljacic J. Meier I, et al. Arabidopsis Book. 2010;8:e0139. doi: 10.1199/tab.0139. Epub 2010 Oct 7. Arabidopsis Book. 2010. PMID: 22303264 Free PMC article. - Ion channels at the nucleus: electrophysiology meets the genome.
Matzke AJ, Weiger TM, Matzke M. Matzke AJ, et al. Mol Plant. 2010 Jul;3(4):642-52. doi: 10.1093/mp/ssq013. Epub 2010 Apr 21. Mol Plant. 2010. PMID: 20410254 Free PMC article. Review. - The temperature-sensitive brush mutant of the legume Lotus japonicus reveals a link between root development and nodule infection by rhizobia.
Maekawa-Yoshikawa M, Müller J, Takeda N, Maekawa T, Sato S, Tabata S, Perry J, Wang TL, Groth M, Brachmann A, Parniske M. Maekawa-Yoshikawa M, et al. Plant Physiol. 2009 Apr;149(4):1785-96. doi: 10.1104/pp.108.135160. Epub 2009 Jan 28. Plant Physiol. 2009. PMID: 19176723 Free PMC article. - Getting to the roots of it: Genetic and hormonal control of root architecture.
Jung JK, McCouch S. Jung JK, et al. Front Plant Sci. 2013 Jun 18;4:186. doi: 10.3389/fpls.2013.00186. eCollection 2013. Front Plant Sci. 2013. PMID: 23785372 Free PMC article. - Buffering capacity explains signal variation in symbiotic calcium oscillations.
Granqvist E, Wysham D, Hazledine S, Kozlowski W, Sun J, Charpentier M, Martins TV, Haleux P, Tsaneva-Atanasova K, Downie JA, Oldroyd GE, Morris RJ. Granqvist E, et al. Plant Physiol. 2012 Dec;160(4):2300-10. doi: 10.1104/pp.112.205682. Epub 2012 Oct 1. Plant Physiol. 2012. PMID: 23027664 Free PMC article.
References
- Harrison, M. J. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 361-389. - PubMed
- Parniske, M. (2000) Current Opin. Plant Biol. 3, 320-328. - PubMed
- Stougaard, J. (2001) Curr. Opin. Plant Biol. 4, 328-335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous