The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation - PubMed (original) (raw)
The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation
Sundeep Kalantry et al. Nat Cell Biol. 2006 Feb.
Abstract
The Polycomb group (PcG) encodes an evolutionarily conserved set of chromatin-modifying proteins that are thought to maintain cellular transcriptional memory by stably silencing gene expression. In mouse embryos that are mutated for the PcG protein Eed, X-chromosome inactivation (XCI) is not stably maintained in extra-embryonic tissues. Eed is a component of a histone-methyltransferase complex that is thought to contribute to stable silencing in undifferentiated cells due to its enrichment on the inactive X-chromosome in cells of the early mouse embryo and in stem cells of the extra-embryonic trophectoderm lineage. Here, we demonstrate that the inactive X-chromosome in Eed(-/-) trophoblast stem cells and in cells of the trophectoderm-derived extra-embryonic ectoderm in Eed(-/-) embryos remain transcriptionally silent, despite lacking the PcG-mediated histone modifications that normally characterize the facultative heterochromatin of the inactive X-chromosome. Whereas undifferentiated Eed(-/-) trophoblast stem cells maintained XCI, reactivation of the inactive X-chromosome occurred when these cells were differentiated. These results indicate that PcG complexes are not necessary to maintain transcriptional silencing of the inactive X-chromosome in undifferentiated stem cells. Instead, PcG proteins seem to propagate cellular memory by preventing transcriptional activation of facultative heterochromatin during differentiation.
Figures
Figure 1
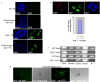
The paternal X-chromosome (Xp) is active only in differentiating _Eed_−/− trophoblast stem (TS) cells. An Xp-linked GFP transgene is used as a reporter of X-linked gene activity and nuclei are stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI). (a) Wild-type (WT) TS cells undergo imprinted XCI of the Xp, as indicated by a complete lack of Xp-GFP expression in all cells, including after differentiation. _Eed_−/− TS colonies contain cells with an active Xp located preferentially at the periphery of the colonies, where differentiated cells are found. (b) Immunofluorescence (IF) detection of Cdx2, a marker of undifferentiated trophectoderm cells, in cultured _Eed_−/− TS cells. Cdx2 is downregulated in mutant cells harboring an active Xp, as indicated by GFP expression, indicating that these cells are differentiated. (c) RT-PCR analysis of male and female WT and _Eed_−/− TS cells for markers of undifferentiated and differentiated trophoblast cells. Cdx2, Eomes (Eomesodermin), and Fgf receptor 2 (FgfR2), all markers of undifferentiated trophectoderm cells, are expressed in all four cell lines examined. Hand1, a marker of intermediate, non-giant differentiated trophoblast cells is also expressed in all cell lines. Pl1 (Placental lactogen 1), a marker of trophoblast giant cells, is absent only in female _Eed_−/− TS cells. Thus, _Eed_−/− female, but not male, TS cells are blocked from terminal differentiation into giant cells, consistent with reactivation of the Xp during initial differentiation of the female mutant TS cells resulting in a block to their further differentiation. (d) Trophoblast (TB) cells located on the periphery of WT blastocyst outgrowths lack Xp-activity, as indicated by Xp-GFP expression, while TB cells in _Eed_−/− female blastocyst outgrowths harbor an active Xp.
Figure 2
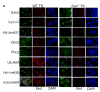
All features of the Xi-heterochromatin are absent in _Eed_−/− female TS cells. (a) IF detection of the Polycomb repressive complex 2 (PRC2) proteins Ezh2, Su(z)12, the PRC2-mediated histone modification tri-methyl lysine 27 of histone H3 (H3-3mK27), the PRC1 proteins Cbx2 and Phc2, PRC1-like mediated histone modification ubiquitylated-H2A (Ub-H2A), Pr-Set7 mediated histone modification mono-methyl lysine 20 of histone H4 (H4-1mK20), and the histone variant macroH2A. Left three rows, WT female TS cells; right three rows, _Eed_−/− female TS cells. Middle panels in both WT and _Eed_−/− TS cell columns are Eed immunostains; right panels are nuclei stained with DAPI. All proteins or epigenetic marks that colocalize with Eed on the inactive-X in WT TS cells, and are absent in all _Eed_−/− TS cells.
Figure 3
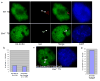
Xist RNA fails to coat the Xp in all _Eed_−/− TS cells. (a) IF-FISH detection of Eed (purple) and Xist RNA (red) in wild-type (WT) and _Eed_−/− TS cells. In WT TS cells, Xist (red) and Eed (purple) colocalize on the Xi in the nucleus (blue); _Eed_−/− TS cells lack Xist RNA coating of the Xi. (b) Trophoblast (TB) cells in cultured _Eed_−/− blastocysts also show lack of Xist RNA accumulation onto the Xi. WT blastocyst outgrowths harbor TB giant cells characterized by larger nuclei with endoreduplicated genomes and multiple inactive-Xs, as marked by multiple Xist foci. _Eed_−/− female embryos do not differentiate TB giant cells, due to X-inactivation defect in diploid TB cells. Inner cell mass-derived cells in _Eed_−/− blastocysts, however, do display Xist RNA accumlation onto the Xi. (c) RT-PCR analysis of Xist and Tsix RNAs in WT and _Eed_−/− TS cells. Xist is expressed in _Eed_−/− TS cells, but its steady-state levels are decreased compared to WT TS cells. Tsix is not detectable in both WT and _Eed_−/− TS cells. (d) Real-time RT-PCR quantitation of Xist RNA in _Eed_−/− TS cells relative to WT TS cells. Male mouse embryonic fibroblast cells serve as control not expressing Xist RNA.
Figure 4
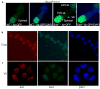
Absence of an epigenetic hallmark of active chromatin, histone H3-di-methyl lysine 4 (H3-2mK4), from the paternal X-chromosome (Xp) in _Eed_−/− TS cells. (a) IF detection of H3-2mK4 (green) and FISH detection of Xist RNA (red) and Xist merged with H3-2mK4 in a representative WT female TS cell nucleus. The Xi, as marked by Xist RNA accumulation, is devoid of H3-2mK4 in all WT TS cells. Xist RNA does not coat the Xi in _Eed_−/− female TS cells but is detected as a pinpoint. In most mutant TS cells, the pinpoint Xist RNA signal falls within a hole devoid of H3-2mK4. Nuclei are stained blue with DAPI. (b) Similar percentages of cells have an active Xp, as assayed by Xp-GFP expression, and an Xp that overlaps with H3-2mK4 staining, suggesting that trophoblast cells with an active Xp also harbor H3-2mK4 on that chromosome. (c) H3-2mK4 IF and FISH detection of the Xp-GFP RNA in _Eed_−/− female TS cells. (d) 95% of Xp-GFP expressing _Eed_−/− TS cells also have H3-2mK4 staining of the Xp. Only differentiating _Eed_−/− TS cells reactivate the Xp (Fig. 1) and these cells no longer exclude marks correlated with transcriptional activity.
Figure 5
Lack of XCI defects in the trophectoderm-derived undifferentiated extra-embryonic ectoderm and the differentiated derivatives of the primitive endoderm in _Eed_−/− embryos. (a) WT embryos do not display any Xp-activity in the extra-embryonic tissues of the embryo, due to imprinted XCI of the Xp in these cells. The epiblast (embryo proper, red arrow) undergoes random XCI, resulting in a mosaicism of X-chromosome activity; in some cells the maternal-X is active and in some cells the Xp is active, as indicated by Xp-GFP expression. E6.2 _Eed_−/− female embryos reactivate the Xp only in differentiating trophoblast (TB) cells in the ectoplacental cone (EPC; the proximal end of the embryo, yellow arrow). Undifferentiated extra-embryonic ectoderm (ExE, white arrow), a source of trophoblast stem (TS) cells and precursors of differentiated trophoblast cells, and visceral endoderm layer (VE, light blue arrow) are devoid of Xp-activity in _Eed_−/− embryos. (b) Absence of PRC2 enrichment on the inactive-Xp in female primitive endoderm-derived (Endo) cell lines. (c) Visceral endoderm cells isolated from E6.5 mouse embryos also accumulate Xist RNA but not Eed on the inactive-X (Xi). Nuclei are stained with DAPI.
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Similar articles
- The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation.
Kalantry S, Magnuson T. Kalantry S, et al. PLoS Genet. 2006 May;2(5):e66. doi: 10.1371/journal.pgen.0020066. Epub 2006 May 5. PLoS Genet. 2006. PMID: 16680199 Free PMC article. - Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes.
Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Silva J, et al. Dev Cell. 2003 Apr;4(4):481-95. doi: 10.1016/s1534-5807(03)00068-6. Dev Cell. 2003. PMID: 12689588 - Developmental regulation of Suz 12 localization.
de la Cruz CC, Fang J, Plath K, Worringer KA, Nusinow DA, Zhang Y, Panning B. de la Cruz CC, et al. Chromosoma. 2005 Aug;114(3):183-92. doi: 10.1007/s00412-005-0008-6. Epub 2005 Jun 29. Chromosoma. 2005. PMID: 15986205 - SETting the stage. Eed-Enx1 leaves an epigenetic signature on the inactive X chromosome.
Chadwick BP, Willard HF. Chadwick BP, et al. Dev Cell. 2003 Apr;4(4):445-7. doi: 10.1016/s1534-5807(03)00097-2. Dev Cell. 2003. PMID: 12689584 Review. - The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome.
Dixon-McDougall T, Brown C. Dixon-McDougall T, et al. Biochem Cell Biol. 2016 Feb;94(1):56-70. doi: 10.1139/bcb-2015-0016. Epub 2015 Jun 24. Biochem Cell Biol. 2016. PMID: 26283003 Review.
Cited by
- The many faces of Polycomb regulation by RNA.
Almeida M, Bowness JS, Brockdorff N. Almeida M, et al. Curr Opin Genet Dev. 2020 Apr;61:53-61. doi: 10.1016/j.gde.2020.02.023. Epub 2020 May 11. Curr Opin Genet Dev. 2020. PMID: 32403014 Free PMC article. Review. - Diverse factors are involved in maintaining X chromosome inactivation.
Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z. Chan KM, et al. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16699-704. doi: 10.1073/pnas.1107616108. Epub 2011 Sep 21. Proc Natl Acad Sci U S A. 2011. PMID: 21940502 Free PMC article. - Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass.
Williams LH, Kalantry S, Starmer J, Magnuson T. Williams LH, et al. Development. 2011 May;138(10):2049-57. doi: 10.1242/dev.061176. Epub 2011 Apr 6. Development. 2011. PMID: 21471155 Free PMC article. - Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation.
Maclary E, Buttigieg E, Hinten M, Gayen S, Harris C, Sarkar MK, Purushothaman S, Kalantry S. Maclary E, et al. Nat Commun. 2014 Jun 30;5:4209. doi: 10.1038/ncomms5209. Nat Commun. 2014. PMID: 24979243 Free PMC article. - H3K27me3-mediated epigenetic regulation in pluripotency maintenance and lineage differentiation.
Jiang L, Huang L, Jiang W. Jiang L, et al. Cell Insight. 2024 Jun 27;3(4):100180. doi: 10.1016/j.cellin.2024.100180. eCollection 2024 Aug. Cell Insight. 2024. PMID: 39072246 Free PMC article. Review.
References
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. - PubMed
- Wang J, et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–5. - PubMed
- Mak W, et al. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr Biol. 2002;12:1016–20. - PubMed
- Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–95. - PubMed
- Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases