Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression - PubMed (original) (raw)
Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression
Qun Pan et al. Genes Dev. 2006.
Abstract
Sequence-based analyses have predicted that approximately 35% of mammalian alternative splicing (AS) events produce premature termination codon (PTC)-containing splice variants that are targeted by the process of nonsense-mediated mRNA decay (NMD). This led to speculation that AS may often regulate gene expression by activating NMD. Using AS microarrays, we show that PTC-containing splice variants are generally produced at uniformly low levels across diverse mammalian cells and tissues, independently of the action of NMD. Our results suggest that most PTC-introducing AS events are not under positive selection pressure and therefore may not contribute important functional roles.
Figures
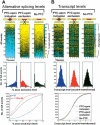
Figure 1.
Microarray comparison of AS and transcript levels of genes containing AS events predicted, based on sequence analysis, to introduce a PTC upon exon inclusion (“PTC-upon inclusion”), introduce a premature termination codon upon exon exclusion (“PTC-upon exclusion”), or not to introduce a PTC (“No PTC”) (refer to Supplementary Tables S1, S2 for more information). Alternative splicing and corresponding transcript levels of genes were profiled in 10 diverse mouse tissues, arranged from left to right in each panel (lanes _1_-10: brain, heart, intestine, kidney, liver, lung, muscle, salivary, spleen, and testis). (A) Alternative splicing levels are represented by percent exon exclusion (i.e., the percent of the total transcripts from a gene with a skipped exon, indicated by the color scale). The AS events are ordered from low to high percent exon exclusion (based on the average value for the 10 tissues) down the _Y_-axis. (B) Transcript levels are represented by the averages of arcsinh-transformed intensity measurements of the probes specific for the flanking constant exons (indicated by the color scale). The order of AS events down the _Y_-axis in B is the same as in A, allowing a direct comparison. Histograms below the microarray clustergrams in A represent the distribution of AS events with the percent exon exclusion level range indicated. Histograms in B represent the distribution of AS events with the transcript levels indicated. The percent exon exclusion and transcript levels measured in the 10 tissues in each of the PTC-upon inclusion, PTC-upon exclusion, and No PTC categories were resampled 10,000 times to normalize for the different numbers of AS events in each category. The line curves (bottom row) show cumulative distributions of percent exon exclusion or transcript levels of the AS events, using the same data as used to plot the histograms. The distributions of percent exon exclusion levels between the PTC-upon inclusion (blue text)/exclusion (red text) and No PTC categories are significantly different (Wilcoxon rank sum test). In contrast, differences in the distributions of transcript levels between the PTC-upon inclusion/exclusion and No PTC categories are not statistically significant.
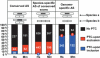
Figure 2.
Microarray analysis of the role of the essential NMD factor UPF1 in controlling AS and transcript levels. (A) Western blot of total cellular protein isolated from HeLa cells 3 d after transfection with either an siRNA that reduces expression of UPF1 (UPF1 kd), or a nonspecific control siRNA (Control) (Kim et al. 2005). The protein lysates were probed with an antibody specific for UPF1 (Lykke-Andersen et al. 2000), and with an anti-Calnexin antibody as a loading control. Serial threefold dilutions of protein in the left three lanes indicate that the Western blot is semiquantitative and permitted an estimation of the UPF1 depletion efficiency at ∼95%. (B) Microarray comparison of percent exon exclusion levels profiled in Control and UPF1-depleted HeLa cells. Genes are ordered down the _Y_-axis from low to high percent exon exclusion in the Control columns, and the same gene orders are preserved in the UPF1 kd columns. Data for percent exon exclusion levels are shown for the three AS event categories (PTC-upon inclusion/exclusion and No PTC). The color scale is as shown in Figure 1A. (C) The bar graphs indicate the percent of AS events (_Y_-axis) in the three AS event categories that show an increase, decrease, or no change in percent exon exclusion level, measured in each case as the difference in the percent exon exclusion between the UPF1 kd and Control kd. Different cutoffs (ranging from at least 5% to at least 15%) for the difference in percent exon exclusion levels were used, as indicated on the _X_-axis. The enrichment for percent exon exclusion changes in the PTC-upon inclusion/exclusion categories that result in increased levels of the PTC-containing variant upon UPF1 knockdown is statistically significant (p < 10-4 for 5%-10% difference and p < 0.05 for 15% difference, Fisher's exact test). (D) The difference in percent exon exclusion values (upper panel) and ratios of transcript levels (lower panel) between the UPF1-depleted and Control samples for the three AS event categories are displayed in cumulative distribution plots. These plots were generated using data for the same AS genes as shown in B, which represent all AS events with GenASAP percent exon exclusion values ranking in the top half of the data (refer to the text). The microarray measurements in the PTC-upon inclusion/exclusion and No PTC were resampled 10,000 times to normalize for different numbers of AS events in each category. The changes in percent exon exclusion levels between the PTC-upon inclusion/exclusion and No PTC categories, upon UPF1 knockdown, are significantly different (Wilcoxon rank sum test). The differences in ratios of transcript levels between the PTC-upon inclusion/exclusion and No PTC categories, upon UPF1 knockdown, are not statistically significant.
Figure 3.
AS events with the potential to trigger NMD are more often represented by species-specific alternative exons than conserved alternative exons. Percentages of total AS events with or without the potential to activate NMD that are conserved or species-specific in human and mouse are shown. Alternative exons were identified in human and mouse ortholog gene pairs using EST and cDNA sequence data, as described previously (Pan et al. 2005) (see also Materials and Methods), and also scored for their potential to introduce a PTC and elicit NMD. “Conserved AS” indicates detection of sequence-conserved alternative and flanking exons in a human and mouse ortholog pair; “Species-specific AS of conserved exons” indicates a conserved exon that is alternatively spliced in one species and constitutively spliced in the other; and “Genome-specific AS” indicates detection of an AS event in one species and sequence evidence for only the spliced constitutive exons in the other species.
Similar articles
- Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay.
Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Saltzman AL, et al. Mol Cell Biol. 2008 Jul;28(13):4320-30. doi: 10.1128/MCB.00361-08. Epub 2008 Apr 28. Mol Cell Biol. 2008. PMID: 18443041 Free PMC article. - Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay.
McGlincy NJ, Tan LY, Paul N, Zavolan M, Lilley KS, Smith CW. McGlincy NJ, et al. BMC Genomics. 2010 Oct 14;11:565. doi: 10.1186/1471-2164-11-565. BMC Genomics. 2010. PMID: 20946641 Free PMC article. - Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay.
Silva AL, Ribeiro P, Inácio A, Liebhaber SA, Romão L. Silva AL, et al. RNA. 2008 Mar;14(3):563-76. doi: 10.1261/rna.815108. Epub 2008 Jan 29. RNA. 2008. PMID: 18230761 Free PMC article. - The coupling of alternative splicing and nonsense-mediated mRNA decay.
Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. Lareau LF, et al. Adv Exp Med Biol. 2007;623:190-211. doi: 10.1007/978-0-387-77374-2_12. Adv Exp Med Biol. 2007. PMID: 18380348 Review. - The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision?
Silva AL, Romão L. Silva AL, et al. FEBS Lett. 2009 Feb 4;583(3):499-505. doi: 10.1016/j.febslet.2008.12.058. Epub 2009 Jan 20. FEBS Lett. 2009. PMID: 19162024 Review.
Cited by
- Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT).
Biamonti G, Bonomi S, Gallo S, Ghigna C. Biamonti G, et al. Cell Mol Life Sci. 2012 Aug;69(15):2515-26. doi: 10.1007/s00018-012-0931-7. Epub 2012 Feb 19. Cell Mol Life Sci. 2012. PMID: 22349259 Free PMC article. Review. - Post-transcriptional regulation of 5-lipoxygenase mRNA expression via alternative splicing and nonsense-mediated mRNA decay.
Ochs MJ, Sorg BL, Pufahl L, Grez M, Suess B, Steinhilber D. Ochs MJ, et al. PLoS One. 2012;7(2):e31363. doi: 10.1371/journal.pone.0031363. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363630 Free PMC article. - Navigating without a road map.
Culbertson MR. Culbertson MR. Genetics. 2007 Sep;177(1):1-7. doi: 10.1093/genetics/177.1.1. Genetics. 2007. PMID: 17890360 Free PMC article. No abstract available. - Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site.
Dou Y, Fox-Walsh KL, Baldi PF, Hertel KJ. Dou Y, et al. RNA. 2006 Dec;12(12):2047-56. doi: 10.1261/rna.151106. Epub 2006 Oct 19. RNA. 2006. PMID: 17053087 Free PMC article. - Global analysis of mRNA splicing.
Moore MJ, Silver PA. Moore MJ, et al. RNA. 2008 Feb;14(2):197-203. doi: 10.1261/rna.868008. Epub 2007 Dec 14. RNA. 2008. PMID: 18083834 Free PMC article. Review.
References
- Alonso, C.R. 2005. Nonsense-mediated RNA decay: A molecular system micromanaging individual gene activities and suppressing genomic noise. Bioessays 27: 463-466. - PubMed
- Baker, K.E. and Parker, R. 2004. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16: 293-299. - PubMed
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291-336. - PubMed
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials