Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region - PubMed (original) (raw)
Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region
Pierre Therizols et al. J Cell Biol. 2006.
Erratum in
- J Cell Biol. 2006 Mar 13;172(6):951
Abstract
In the yeast Saccharomyces cerevisiae that lacks lamins, the nuclear pore complex (NPC) has been proposed to serve a role in chromatin organization. Here, using fluorescence microscopy in living cells, we show that nuclear pore proteins of the Nup84 core complex, Nup84p, Nup145Cp, Nup120p, and Nup133p, serve to anchor telomere XI-L at the nuclear periphery. The integrity of this complex is shown to be required for repression of a URA3 gene inserted in the subtelomeric region of this chromosome end. Furthermore, altering the integrity of this complex decreases the efficiency of repair of a DNA double-strand break (DSB) only when it is generated in the subtelomeric region, even though the repair machinery is functional. These effects are specific to the Nup84 complex. Our observations thus confirm and extend the role played by the NPC, through the Nup84 complex, in the functional organization of chromatin. They also indicate that anchoring of telomeres is essential for efficient repair of DSBs occurring therein and is important for preserving genome integrity.
Figures
Figure 1.
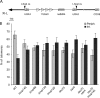
Nup84 complex is required for tethering of tel-XIL at the periphery. (A) Map of _tet_O integration site on chromosome XI-L. One _tet_O tract of 112 repeats is integrated 3.5 kb from the telomere of left arm of chromosome XI (XI-L). Gray rectangles, genes; open triangles, integrated _tet_O sequences; heavy black line, chromosome; thin black line, plasmid sequence; horizontal black arrow, telomere; vertical black arrows, I-SceI cutting sites. Scale in kilobases (kb) is indicated. COS9 (YKL219w) is located 14.5 kb from the telomere; double slash indicates an interruption in the chromosome representation. (B) Proportion of telomeres found in each volume of the nucleus in the different genetic backgrounds. Standard errors with a P value of 0.05 are shown. Light gray columns represent values observed in peripheral volume, dark gray columns in the internal one.
Figure 2.
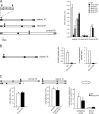
Nup84 complex is required for subtelomeric DSB repair efficiencies. (A) Survival to DSBs according to their position in wild-type and different nucleoporin mutant backgrounds. Chromosomes XI and XV are schematized. Black dots and vertical hatched boxes represent centromere and subtelomeres, respectively. Arrows indicate the positions of induced DSBs generated by cleavage of two inverted I-SceI cutting sites (cs) surrounding the URA3 marker (black box). Size in kb is indicated. Colony forming units (CFU) are calculated as the ratio of galactose growing colonies (GAL) versus glucose growing colonies (GLU) expressed in percentages (%). Values obtained for wild-type (WT, white), Δnup145C (black), Δnup84 (gray), Δnup133 (diagonals), Δnup120 (horizontal lines), and Δnup170 (white dots) are shown according to DSB's position. Subtel. XI stands for subtelomeric XI-L. Standard errors with a P value of 0.1 are shown. (B) Proportion of repair by NHEJ and rearrangements in subtelomere XI-L. Subtelomeric DSB is schematized as in A. Percentages of repair either by NHEJ or by gross chromosomal rearrangements per plated cells are shown for wild-type (WT, white) and Δnup145C (black) (C). Repair machinery is functional in Δnup145C. Left; DSB is generated in a central part of chromosome XV and gene conversion is made possible because of homologous sequences present on chromosome V (gray box, see text and Karathanasis and Wilson, 2002). Colony forming units are calculated as in A. Middle; DSB is generated in the subtelomeric position in diploid wild-type cells or diploid cells homozygous for Δnup145C. Colony forming units are calculated as in A. Right; DSB is created on a replicative plasmid by restriction enzymes generating cohesive or blunt ended ends. Wild-type (WT) and Δnup145C strains were transformed with equivalent amounts of supercoiled or linearized pRS316 (ARS-CEN-URA3) plasmid. The number of transformants obtained with the linearized plasmid expressed as a percentage of the number of transformants obtained with the supercoil DNA are plotted. Experiments were repeated three times and error bars represent the 95% confidence intervals.
Figure 3.
Sequences of the junction of NHEJ events at locus L1 in Δnup145C and proposed mechanisms of NHEJ. Coordinates of the L1 locus on chromosome XI-L are shown. I-SceI restriction sites (in opposite orientations) are in bold, cleavage position generating 4-bp 3′ overhangs is indicated by a staggered line, BamHI and KpnI restriction sites are boxed and plasmid sequences are in lower case letters (see Fairhead et al., 1996 for construction details). Possible end processing and subsequent alignment of complementary base pairs are shown. See text for details.
Figure 4.
Mutants of the Nup84 complex are defective for subtelomeric silencing. (A) The native configuration of chromosome XI-L subtelomere is schematized. Black arrows represent TG1-3 repeats; gray box, the coreX sequence which includes the ACS sequence; white triangle, the position of URA3 insertion (see Pryde and Louis, 1999 for details). (B) Measurement of repression of URA3 inserted at ACS of coreX. 10-fold dilutions were spotted on rich medium (YPD), on synthetic complete medium lacking uracil (SC-URA), and on medium containing 5-FOA. On this last medium wild-type cells bearing URA3 inserted into ACS sequence of core X (URA3@X-ACS) are able to repress this gene as illustrated by their ability to grow on FOA-containing medium. On the contrary, minimal repression is seen in Nup84 complex mutants (Δnup145C, Δnup84, Δnup120, Δnup133). No effect on telomeric repression is seen in other mutants (Δnup170, Δnup53). Silencing defects are complemented by full-length (FL) or the carboxy-terminal encoding part (C) of corresponding wild-type gene; Δnup133 silencing defect are complemented both by the wild-type gene (FL) or the nup133ΔN separation-of-function mutant (ΔN) that clusters NPCs (see Doye et al., 1994 for details). (C) Quantification of silencing defects. Cells were grown in nonselective conditions and 10-fold dilutions were plated onto YPD and medium containing 5-FOA.
Figure 5.
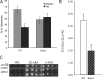
Δesc1 is defective for telomere tethering and subtelomeric DSB repair. (A) The proportion of tagged telomere XI-L in nuclear space was quantified as in Fig. 1 B. Light gray, peripheral volume; dark gray, internal volume. Bars represent the 95% confidence interval. (B) Survival to subtelomeric DSB is represented as colony forming units (CFU) as in Fig. 2 A and expressed in percentages (%). Values obtained for wild-type (WT, white) and Δesc1 (checkered) are shown. (C) Measurement of repression of URA3 inserted at ACS of coreX of telomere XI-L as in Fig. 4. 10-fold dilutions were spotted on rich medium (YPD), on synthetic complete medium lacking uracil (SC-URA), and on medium containing 5-FOA.
Similar articles
- Absence of yKu/Hdf1 but not myosin-like proteins alters chromosome dynamics during prophase I in yeast.
Scherthan H, Trelles-Sticken E. Scherthan H, et al. Differentiation. 2008 Jan;76(1):91-8. doi: 10.1111/j.1432-0436.2007.00212.x. Epub 2007 Aug 14. Differentiation. 2008. PMID: 17697124 - Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast.
Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U. Feuerbach F, et al. Nat Cell Biol. 2002 Mar;4(3):214-21. doi: 10.1038/ncb756. Nat Cell Biol. 2002. PMID: 11862215 - Repairing subtelomeric DSBs at the nuclear periphery.
Taddei A, Gasser SM. Taddei A, et al. Trends Cell Biol. 2006 May;16(5):225-8. doi: 10.1016/j.tcb.2006.03.005. Epub 2006 Apr 18. Trends Cell Biol. 2006. PMID: 16621562 Review. - A study of biochemical and functional interactions of Htl1p, a putative component of the Saccharomyces cerevisiae, Rsc chromatin-remodeling complex.
Florio C, Moscariello M, Ederle S, Fasano R, Lanzuolo C, Pulitzer JF. Florio C, et al. Gene. 2007 Jun 15;395(1-2):72-85. doi: 10.1016/j.gene.2007.02.002. Epub 2007 Feb 20. Gene. 2007. PMID: 17400406 - Telomeres and the DNA damage response: why the fox is guarding the henhouse.
Maser RS, DePinho RA. Maser RS, et al. DNA Repair (Amst). 2004 Aug-Sep;3(8-9):979-88. doi: 10.1016/j.dnarep.2004.05.009. DNA Repair (Amst). 2004. PMID: 15279784 Review.
Cited by
- The polyHIS Tract of Yeast AMPK Coordinates Carbon Metabolism with Iron Availability.
Simpson-Lavy KJ, Kupiec M. Simpson-Lavy KJ, et al. Int J Mol Sci. 2023 Jan 10;24(2):1368. doi: 10.3390/ijms24021368. Int J Mol Sci. 2023. PMID: 36674878 Free PMC article. - The nuclear pore complex: bridging nuclear transport and gene regulation.
Strambio-De-Castillia C, Niepel M, Rout MP. Strambio-De-Castillia C, et al. Nat Rev Mol Cell Biol. 2010 Jul;11(7):490-501. doi: 10.1038/nrm2928. Nat Rev Mol Cell Biol. 2010. PMID: 20571586 Review. - Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres.
Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Therizols P, et al. Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):2025-30. doi: 10.1073/pnas.0914187107. Epub 2010 Jan 13. Proc Natl Acad Sci U S A. 2010. PMID: 20080699 Free PMC article. - Components and regulation of nuclear transport processes.
Cautain B, Hill R, de Pedro N, Link W. Cautain B, et al. FEBS J. 2015 Feb;282(3):445-62. doi: 10.1111/febs.13163. Epub 2014 Dec 22. FEBS J. 2015. PMID: 25429850 Free PMC article. Review. - Biology and biophysics of the nuclear pore complex and its components.
Lim RY, Ullman KS, Fahrenkrog B. Lim RY, et al. Int Rev Cell Mol Biol. 2008;267:299-342. doi: 10.1016/S1937-6448(08)00632-1. Int Rev Cell Mol Biol. 2008. PMID: 18544502 Free PMC article. Review.
References
- Andrulis, E.D., A.M. Neiman, D.C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 394:592–595. - PubMed
- Aten, J.A., J. Stap, P.M. Krawczyk, C.H. van Oven, R.A. Hoebe, J. Essers, and R. Kanaar. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 303:92–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases