Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli - PubMed (original) (raw)
Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli
Scott J Weissman et al. Mol Microbiol. 2006 Feb.
Abstract
Type 1 fimbriae of Escherichia coli mediate mannose-specific adhesion to host epithelial surfaces and consist of a major, antigenically variable pilin subunit, FimA, and a minor, structurally conserved adhesive subunit, FimH, located on the fimbrial tip. We have analysed the variability of fimA and fimH in strains of vaginal and other origin that belong to one of the most prominent clonal groups of extraintestinal pathogenic E. coli, comprised of O1:K1-, O2:K1- and O18:K1-based serotypes. Multiple locus sequence typing (MLST) of this group revealed that the strains have identical (at all but one nucleotide position) eight housekeeping loci around the genome and belong to the ST95 complex defined by the publicly available E. coli MLST database. Multiple highly diverse fimA alleles have been introduced into the ST95 clonal complex via horizontal transfer, at a frequency comparable to that of genes defining the major O- and H-antigens. However, no further significant FimA diversification has occurred via point mutation after the transfers. In contrast, while fimH alleles also move horizontally (along with the fimA loci), they acquire point amino acid replacements at a higher rate than either housekeeping genes or fimA. These FimH mutations enhance binding to monomannose receptors and bacterial tropism for human vaginal epithelium. A similar pattern of rapid within-clonal structural evolution of the adhesive, but not pilin, subunit is also seen, respectively, in papG and papA alleles of the di-galactose-specific P-fimbriae. Thus, while structurally diverse pilin subunits of E. coli fimbriae are under selective pressure for frequent horizontal transfer between clones, the adhesive subunits of extraintestinal E. coli are under strong positive selection (Dn/Ds > 1 for fimH and papG) for functionally adaptive amino acid replacements.
Figures
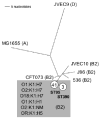
Fig. 1
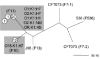
Unrooted phylogram of MLST loci of E. coli K1 and non-K1 archetypal strains. Based on concatenated 500 bp fragments of MLST housekeeping loci adk, fumC, gyrB, icd, mdh, metG, purA and recA. Branch lengths reflect absolute nucleotide differences between concatenated sequences. Circled numbers indicate number of strains at specified node. Letters in parentheses indicate major phylogenetic group assignment. Grey box indicates serotypes belonging to ST95 and ST390. CFT073, 536 and J96 are archetypal extraintestinal pathogenic E. coli strains. MG1655 and JVEC9 represent out-group strains.
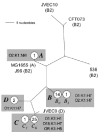
Fig. 2
Unrooted phylogram of fimA sequences from K1 and non-K1 archetypal strains. Labelling conventions same as for Fig. 1, with the following additions. Non-ST95 strains with sequences identical to complex strains are placed next to, but outside of, grey boxes, without branches. Letters in italics (with and without subscripts) indicate alleles as discussed in text.
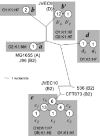
Fig. 3
Unrooted phylogram of fimH alleles from K1 and non-K1 archetypal strains. Figure conventions same as for Figs 1 and 2.
Fig. 4
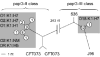
CEL-1 digests of homo- and heteroduplexes of fim clusters cloned from three E. coli O18:K1:H7 strains with distinct fimH alleles. A. fim clusters prior to CEL-1 digestion. Lanes 1–3: homoduplexes of F3, RS218 and 89–1449; lanes 4–6: heteroduplexes of 89–1449/F3, RS218/F3 and 89–1449/RS218. B. fim clusters after CEL-I digestion (lane numbering same as for A). White arrowheads indicate fragments generated by CEL-1 cleavage at heteroduplex SNP sites, with similar pattern observed for all three pairs.
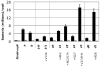
Fig. 5
Monomannose binding properties of distinct fimH alleles expressed in isogenic background. Monomannose binding without inhibitor shown in black bars, with inhibitor in grey. Alleles identified both by letter/subscript terminology used in text and by defining mutations (relative to primary allele in each lineage). SNP-containing allele lineages b 0, c 0 and d 0 demarcated by vertical lines. Error bars indicate standard error of the mean.
Fig. 6
Primary vaginal epithelial cell binding properties of variant fimH alleles expressed in isogenic background, plotted against monomannose binding strength. _R_2 indicates slope of trend line. Error bars indicate standard error of the mean associated with vaginal cell binding data.
Fig. 7
Unrooted phylogram of papA alleles. PapA pilin variants, serologic F-types F10 and F11. Other F-types indicated in parentheses. Circled numbers indicate number of strains at specified node. Branch lengths reflect absolute nucleotide differences between sequences. Grey boxes indicate serotypes belonging to ST95. CFT073, 536 and J96 are archetypal extraintestinal pathogenic E. coli strains. Note that two CFT073 alleles are included.
Fig. 8
Unrooted phylogram of papG alleles. PapG adhesin variants, classes II and III. Figure conventions otherwise same as for Fig. 7.
Similar articles
- Phylogenetic analysis of inflammatory bowel disease associated Escherichia coli and the fimH virulence determinant.
Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Sepehri S, et al. Inflamm Bowel Dis. 2009 Nov;15(11):1737-45. doi: 10.1002/ibd.20966. Inflamm Bowel Dis. 2009. PMID: 19462430 - Sequence analysis demonstrates the conservation of fimH and variability of fimA throughout avian pathogenic Escherichia coli (APEC).
Vandemaele F, Vandekerchove D, Vereecken M, Derijcke J, Dho-Moulin M, Goddeeris BM. Vandemaele F, et al. Vet Res. 2003 Mar-Apr;34(2):153-63. doi: 10.1051/vetres:2002062. Vet Res. 2003. PMID: 12657207 - Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease.
Iebba V, Conte MP, Lepanto MS, Di Nardo G, Santangelo F, Aloi M, Totino V, Checchi MP, Longhi C, Cucchiara S, Schippa S. Iebba V, et al. Infect Immun. 2012 Apr;80(4):1408-17. doi: 10.1128/IAI.06181-11. Epub 2012 Jan 30. Infect Immun. 2012. PMID: 22290143 Free PMC article. - AFA and F17 adhesins produced by pathogenic Escherichia coli strains in domestic animals.
Le Bouguénec C, Bertin Y. Le Bouguénec C, et al. Vet Res. 1999 Mar-Jun;30(2-3):317-42. Vet Res. 1999. PMID: 10367361 Review. - Virulence factors of septicemic Escherichia coli strains.
Mokady D, Gophna U, Ron EZ. Mokady D, et al. Int J Med Microbiol. 2005 Oct;295(6-7):455-62. doi: 10.1016/j.ijmm.2005.07.007. Int J Med Microbiol. 2005. PMID: 16238019 Review.
Cited by
- Differentiation of Crohn's Disease-Associated Isolates from Other Pathogenic Escherichia coli by Fimbrial Adhesion under Shear Force.
Szunerits S, Zagorodko O, Cogez V, Dumych T, Chalopin T, Alvarez Dorta D, Sivignon A, Barnich N, Harduin-Lepers A, Larroulet I, Yanguas Serrano A, Siriwardena A, Pesquera A, Zurutuza A, Gouin SG, Boukherroub R, Bouckaert J. Szunerits S, et al. Biology (Basel). 2016 Apr 1;5(2):14. doi: 10.3390/biology5020014. Biology (Basel). 2016. PMID: 27043645 Free PMC article. - Positive selection identifies an in vivo role for FimH during urinary tract infection in addition to mannose binding.
Chen SL, Hung CS, Pinkner JS, Walker JN, Cusumano CK, Li Z, Bouckaert J, Gordon JI, Hultgren SJ. Chen SL, et al. Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22439-44. doi: 10.1073/pnas.0902179106. Epub 2009 Dec 16. Proc Natl Acad Sci U S A. 2009. PMID: 20018753 Free PMC article. - Type 1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of Escherichia coli.
Tchesnokova V, Aprikian P, Kisiela D, Gowey S, Korotkova N, Thomas W, Sokurenko E. Tchesnokova V, et al. Infect Immun. 2011 Oct;79(10):3895-904. doi: 10.1128/IAI.05169-11. Epub 2011 Jul 18. Infect Immun. 2011. PMID: 21768279 Free PMC article. - Comparative evolutionary analysis of the major structural subunit of Vibrio vulnificus type IV pili.
Chattopadhyay S, Paranjpye RN, Dykhuizen DE, Sokurenko EV, Strom MS. Chattopadhyay S, et al. Mol Biol Evol. 2009 Oct;26(10):2185-96. doi: 10.1093/molbev/msp124. Epub 2009 Jun 25. Mol Biol Evol. 2009. PMID: 19556347 Free PMC article. - Fate of the H-NS-repressed bgl operon in evolution of Escherichia coli.
Sankar TS, Neelakanta G, Sangal V, Plum G, Achtman M, Schnetz K. Sankar TS, et al. PLoS Genet. 2009 Mar;5(3):e1000405. doi: 10.1371/journal.pgen.1000405. Epub 2009 Mar 6. PLoS Genet. 2009. PMID: 19266030 Free PMC article.
References
- Achtman M, Pluschke G. Clonal analysis of descent and virulence among selected Escherichia coli. Annu Rev Microbiol. 1986;40:185–210. - PubMed
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
- Bloch CA, Stocker BA, Orndorff PE. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol. 1992;6:697–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- L40 AI057547/AI/NIAID NIH HHS/United States
- P01 DK053369/DK/NIDDK NIH HHS/United States
- R01 DK070906/DK/NIDDK NIH HHS/United States
- K08 AI057737/AI/NIAID NIH HHS/United States
- R01 GM060731-05/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical