Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease pim1 - PubMed (original) (raw)
Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease pim1
Tamara Major et al. Mol Cell Biol. 2006 Feb.
Abstract
ATP-dependent oligomeric proteases are major components of cellular protein quality control systems. To investigate the role of proteolytic processes in the maintenance of mitochondrial functions, we analyzed the dynamic behavior of the mitochondrial proteome of Saccharomyces cerevisiae by two-dimensional (2D) polyacrylamide gel electrophoresis. By a characterization of the influence of temperature on protein turnover in isolated mitochondria, we were able to define four groups of proteins showing a differential susceptibility to proteolysis. The protein Pim1/LON has been shown to be the main protease in the mitochondrial matrix responsible for the removal of damaged or nonnative proteins. To assess the substrate range of Pim1 under in vivo conditions, we performed a quantitative comparison of the 2D protein spot patterns between wild-type and pim1Delta mitochondria. We were able to identify a novel subset of mitochondrial proteins that are putative endogenous substrates of Pim1. Using an in organello degradation assay, we confirmed the Pim1-specific, ATP-dependent proteolysis of the newly identified substrate proteins. We could demonstrate that the functional integrity of the Pim1 substrate proteins, in particular, the presence of intact prosthetic groups, had a major influence on the susceptibility to proteolysis.
Figures
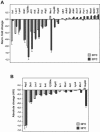
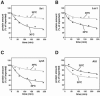
FIG. 1.
Submitochondrial localization and functional classification of the proteins identified from the 2D-PAGE analysis of the mitochondrial proteome. (A and B) Submitochondrial localization of proteins identified in this study (A) and according to the MitoP2 database (B). MA, mitochondrial matrix; IM, inner membrane; IMS, intermembrane space; OM, outer membrane; ND, not defined. (C and D) Functional classes of mitochondrial proteins identified in this study (C) and those included in the MitoP2 database (D). The percentage of total proteins detected within each category is indicated.
FIG. 2.
Temperature-dependent changes in the mitochondrial proteome. (A) Relative changes in protein abundance during a time course of 4 h. Purified mitochondria were incubated at the indicated temperatures under native buffer conditions in the presence of an ATP-regenerating system. The reduction of protein amounts of selected proteins during the incubation period at 30°C and 39°C is represented as normalized (Norm.) change values. As an example, a reduction of 50% would result in a negative value of 1. Error bars indicate the relative error of the change values. (B) Absolute changes in protein abundance. Shown are the differences in spot volumes before and after the incubation period in arbitrary units (AU) obtained from the Imagemaster 2D quantification of the respective protein spots. Error bars indicate the relative errors of the changes in protein amounts.
FIG. 3.
2D-PAGE analysis of the proteome of wild-type and _pim1_Δ mitochondria. Intact isolated mitochondria from wild-type and _pim1_Δ cells were incubated for 1 h at 30°C and analyzed by 2D-PAGE. Protein spots were visualized by Coomassie staining. A typical master 2D map of mitochondrial proteins (wild type) indicating the location of the enlarged sections is shown. Gel sections of WT and _pim1_Δ mitochondria indicate the localization of protein spots with an increased abundance in _pim1_Δ and control proteins (Aco1, Mdh1) (arrows). MW, molecular mass.
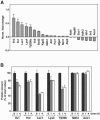
FIG. 4.
Quantification of the differences in mitochondrial protein composition between wild-type and _pim1_Δ mitochondria. (A) Normalized (Norm.) change values for selected proteins of wild-type and _pim1_Δ mutant mitochondria from the experiment in Fig. 3 are shown with gray bars. The proteins are sorted according to degree of change. The relative errors of the change values are represented by error bars. (B) Time course of degradation of selected proteins in wild-type mitochondria. The amount of each protein (spot volume) present at 0 h, after 1 h, and after 4 h of incubation of isolated mitochondria at 30°C was determined. Mitochondrial protein composition was analyzed by 2D-PAGE as described in the text. The protein amount present in the sample at 0 h was set as 100%. The average of three independent experiments is shown. Error bars show the relative errors of the spot volumes.
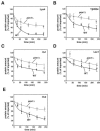
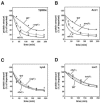
FIG. 5.
Degradation kinetics of selected proteins after import into wild-type and _pim1_Δ mitochondria. The proteolytic breakdown of radiolabeled mitochondrial proteins was monitored at 30°C after the import in wild-type and _pim1_Δ mitochondria. To obtain comparable conditions, the [_rho_0] wild-type strain KRY01 was used. Aliquots were taken and analyzed by SDS-PAGE at various time points (0, 30, 60, 120, 180, and 240 min). The remaining protein amounts in each sample were quantified by autoradiography. The amount of protein present at 0 min was set to 100% for wild-type and _pim1_Δ mitochondria. The diagrams show the means and the standard errors of the means of at least three independent experiments.
FIG. 6.
Stability of control proteins in wild-type and _pim1_Δ mitochondria. Radiolabeled preproteins, Mdh1 (A) and Aco1 (B), were imported into wild-type (WT) and _pim1_Δ mitochondria, and their proteolysis was monitored for 240 min at 30°C. The amount of reporter proteins was quantified as described in the legend to Fig. 5. The means and the standard errors of the means of at least three independent experiments are shown in both diagrams.
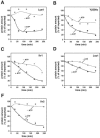
FIG. 7.
ATP-dependent degradation of putative Pim1 substrates. After import of the indicated radiolabeled preproteins, Lys4 (A), Yjl200c (B), Ilv1 (C), Lsc1 (D), and Ilv2 (E), in wild-type mitochondria, the samples were incubated at 30°C for 240 min either with an ATP-regenerating system (+ ATP) or with apyrase and oligomycin (− ATP). The degradation of the imported proteins was analyzed by SDS-PAGE and autoradiography as described in the legend to Fig. 5.
FIG. 8.
Temperature-dependent degradation of Pim1 substrates. The proteins Ilv1 (A), Lsc1 (B), Lys4 (C), and, as a control, Alt1 (D) were imported as radiolabeled precursors for 20 min at 30°C into the matrix of wild-type mitochondria. After completion of import, their stability was assayed for 240 min at 30°C or alternatively at 39°C as described in the legend to Fig. 5.
FIG. 9.
Proteolysis of proteins after import into Fe/S cluster assembly mutants. The degradation of the imported and processed radiolabeled proteins Yjl200c (A), Aco1 (B), Lys4 (C), and control protein Lsc1 (D) was analyzed in wild-type, _ssq1_Δ and _nfu1_Δ mitochondria. The stability of the proteins was examined and quantified as described in the legend to Fig. 5.
Similar articles
- Structural properties of substrate proteins determine their proteolysis by the mitochondrial AAA+ protease Pim1.
von Janowsky B, Knapp K, Major T, Krayl M, Guiard B, Voos W. von Janowsky B, et al. Biol Chem. 2005 Dec;386(12):1307-17. doi: 10.1515/BC.2005.149. Biol Chem. 2005. PMID: 16336126 - Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1.
Bayot A, Gareil M, Rogowska-Wrzesinska A, Roepstorff P, Friguet B, Bulteau AL. Bayot A, et al. J Biol Chem. 2010 Apr 9;285(15):11445-57. doi: 10.1074/jbc.M109.065425. Epub 2010 Feb 11. J Biol Chem. 2010. PMID: 20150421 Free PMC article. - The role of protein quality control in mitochondrial protein homeostasis under oxidative stress.
Bender T, Leidhold C, Ruppert T, Franken S, Voos W. Bender T, et al. Proteomics. 2010 Apr;10(7):1426-43. doi: 10.1002/pmic.200800619. Proteomics. 2010. PMID: 20186747 - ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae.
Van Dyck L, Langer T. Van Dyck L, et al. Cell Mol Life Sci. 1999 Nov 30;56(9-10):825-42. doi: 10.1007/s000180050029. Cell Mol Life Sci. 1999. PMID: 11212342 Free PMC article. Review. - [ATP-dependent proteases in the quality control of mitochondrial proteome].
Kołodziejczak M, Opalińska M, Czarna M, Jańska H. Kołodziejczak M, et al. Postepy Biochem. 2016;62(2):206-215. Postepy Biochem. 2016. PMID: 28132473 Review. Polish.
Cited by
- Mitochondrial protein quality control: the mechanisms guarding mitochondrial health.
Bohovych I, Chan SS, Khalimonchuk O. Bohovych I, et al. Antioxid Redox Signal. 2015 Apr 20;22(12):977-94. doi: 10.1089/ars.2014.6199. Epub 2015 Feb 11. Antioxid Redox Signal. 2015. PMID: 25546710 Free PMC article. Review. - Comparative mitochondrial proteomics: perspective in human diseases.
Jiang Y, Wang X. Jiang Y, et al. J Hematol Oncol. 2012 Mar 18;5:11. doi: 10.1186/1756-8722-5-11. J Hematol Oncol. 2012. PMID: 22424240 Free PMC article. Review. - Mrx6 regulates mitochondrial DNA copy number in Saccharomyces cerevisiae by engaging the evolutionarily conserved Lon protease Pim1.
Göke A, Schrott S, Mizrak A, Belyy V, Osman C, Walter P. Göke A, et al. Mol Biol Cell. 2020 Mar 19;31(7):527-545. doi: 10.1091/mbc.E19-08-0470. Epub 2019 Sep 18. Mol Biol Cell. 2020. PMID: 31532710 Free PMC article. - Feedback Control of a Two-Component Signaling System by an Fe-S-Binding Receiver Domain.
Stein BJ, Fiebig A, Crosson S. Stein BJ, et al. mBio. 2020 Mar 17;11(2):e03383-19. doi: 10.1128/mBio.03383-19. mBio. 2020. PMID: 32184258 Free PMC article. - The Mitochondrial Lon Protease: Novel Functions off the Beaten Track?
Voos W, Pollecker K. Voos W, et al. Biomolecules. 2020 Feb 7;10(2):253. doi: 10.3390/biom10020253. Biomolecules. 2020. PMID: 32046155 Free PMC article. Review.
References
- Augustin, S., M. Nolden, S. Müller, O. Hardt, I. Arnold, and T. Langer. 2005. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J. Biol. Chem. 280:2691-2699. - PubMed
- Bateman, J. M., M. Iacovino, P. S. Perlman, and R. A. Butow. 2002. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. J. Biol. Chem. 277:47946-47953. - PubMed
- Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. - PubMed
- Bota, D. A., and K. J. Davies. 2002. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4:674-680. - PubMed
- Catrein, I., R. Herrmann, A. Bosserhoff, and T. Ruppert. 2005. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 272:2892-2900. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases