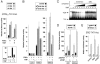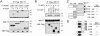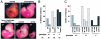TGIF inhibits retinoid signaling - PubMed (original) (raw)
TGIF inhibits retinoid signaling
Laurent Bartholin et al. Mol Cell Biol. 2006 Feb.
Abstract
TGIF (TG-interacting factor) represses transforming growth factor beta (TGF-beta)-activated gene expression and can repress transcription via a specific retinoid response element. Mutations in human TGIF are associated with holoprosencephaly, a severe defect of craniofacial development with both genetic and environmental causes. Both TGF-beta and retinoic acid signaling are implicated in craniofacial development. Here, we analyze the role of TGIF in regulating retinoid responsive gene expression. We demonstrate that TGIF interacts with the ligand binding domain of the RXRalpha retinoid receptor and represses transcription from retinoid response elements. TGIF recruits the general corepressor, CtBP, to RXRalpha, and this recruitment is required for full repression by TGIF. Interaction between TGIF and RXRalpha is reduced by the addition of retinoic acid, consistent with a role for TGIF as an RXRalpha transcriptional corepressor. We created a Tgif null mutation in mice and tested the sensitivity of mutant mice to increased levels of retinoic acid. Tgif mutant embryos are more sensitive to retinoic acid-induced teratogenesis, and retinoid target genes are expressed at a higher level in tissues from Tgif null mice. These results demonstrate an important role for TGIF as a transcriptional corepressor, which regulates developmental signaling by retinoic acid, and raises the possibility that TGIF may repress other RXR-dependent transcriptional responses.
Figures
FIG. 1.
Regulation of RXR-dependent transcription by TGIF. HepG2 cells were transfected with a DR1-TATA-luc luciferase reporter, together with expression vectors encoding RXRα and TGIF, as indicated (−, not used). 9C-RA was added 24 h prior to analysis, as indicated. In panel A, + indicates 10−7 M 9C-RA, and the titration of increasing 9C-RA was 10−9 M, 10−8 M, and 10−7 M. In panel B, increasing amounts of TGIF were cotransfected as indicated (in ng per duplicate). Luciferase activity was assayed 40 h after transfection and is presented, in arbitrary units, as the mean ± standard deviation of results from duplicate transfections.
FIG. 2.
Repression of DR5-mediated transcription by TGIF. HepG2 cells were transfected with DR5-TATA-luc luciferase reporters, together with expression vectors encoding RXRα and TGIF, as indicated. +, present; −, absent. The DR5 reporters contained either 1, 2, or 4 tandem copies of the DR5 element as indicated. (A) AT-RA or 9C-RA was added 24 h prior to analysis, as indicated, to a final concentration of 10−7 M. The boxed inset in panel A shows the DR5-TATA-luc activity in the absence of added retinoic acid. (B) Cells were transfected with a control reporter lacking DR5 elements (TATA-luc) or with reporters containing 1 or 2 DR5 elements, together with RXRα and TGIF expression constructs, as indicated. 9C-RA was added as indicated 24 h prior to lysis. (C) Nuclear extracts from COS1 cells transfected with a TGIF or control vector were incubated with a radiolabeled probe containing a single consensus TGIF site (CTGTCAA). Unlabeled competitor oligonucleotides, either the consensus TGIF site or the DR5 RARE, were added as indicated. Triangles represent a titrations of a 5-, 20-, 80-, 320-, and 530-fold excesses of unlabeled competitor. The TGIF shifted band and free probe are indicated. (D) Cells were transfected with the DR1 or DR5 (two copy) reporters together with a control vector (pSUPER) or one expressing a TGIF-specific hairpin RNA (siTGIF) and treated with 9C-RA, as indicated, 24 h prior to analysis. Three days after transfection, luciferase activity was assayed. (E) Cells were transfected with a control vector, a TGIF expression plasmid, or a TGIF siRNA vector, together with a luciferase reporter in which transcription is activated by an estrogen response element (ERE). Estrogen (E2) was added, as indicated, 24 h prior to analysis.
FIG. 3.
TGIF interacts with RXRα. (A) COS1 cells were transfected with T7 epitope-tagged RARα or RXRα expression constructs, together with a Flag-tagged TGIF or control vector, and cells were treated with 9C-RA for 1 or 16 h (10−7 M) prior to lysis as indicated. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting (WB) for the presence of coprecipitating RARα or RXRα. A portion of the lysates was subjected to analysis by direct Western blotting to monitor protein expression (below). +, present. (B) Cells were transfected as described for panel A, and 9C-RA was added for 1, 2, 4, or 6 h as indicated. Protein complexes were analyzed as described for panel A. (C) COS1 cells either treated for 16 h with 9C-RA (+) or left untreated (−) were lysed by sonication. Protein complexes were precipitated with a RXRα-specific monoclonal antibody and Western blotted with a TGIF-specific antiserum (above). The expression of TGIF and RXRα was monitored in the lysates by direct Western blotting (below). Arrowheads in panel C indicate specific bands; the bar indicates the heavy chain. The positions of molecular mass markers (in kDa) are shown to the left of each panel. IP, immunoprecipitation.
FIG. 4.
TGIF interacts with the RXRα ligand binding domain. (A) COS1 cells were transfected with expression constructs encoding T7 epitope-tagged RXRα deletion constructs, together with a Flag-tagged TGIF or control vector. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting (WB) for the presence of coprecipitating T7-tagged RXRα constructs. (B) COS1 cells were transfected with T7 epitope-tagged ΔAF1 RXRα, together with a series of Flag-tagged TGIF expression constructs. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting for the presence of coprecipitating RXRα. A portion of the lysates was subjected to analysis by direct Western blotting to monitor protein expression (below). (C) The RXRα deletion constructs used in panel A are shown schematically, together with the amino acids present in each construct. TGIF interaction is indicated to the right (+, interaction; −, none). AF1, activation function 1; DBD, DNA binding domain; LBD, ligand binding domain; AF2, activation function 2. (D) The TGIF deletion constructs are shown schematically, together with the amino acids in each and their interaction with RXRα. The two major conserved domains of vertebrate TGIFs are indicated. HD + 20, homeodomain plus 20 amino acid extension; RD2, repression domain 2; IP, immunoprecipitation.
FIG. 5.
TGIF and RXR cocomplexes. (A) COS1 cells were transfected with T7-tagged ΔAF1 RXRα, β, and γ constructs with or without Flag-TGIF. Complexes were isolated on Flag agarose and analyzed for coprecipitating T7-tagged RXRs. (B and C) COS1 cells were transfected with expression plasmids encoding T7-tagged RARα (B) or PPARγ (C), together with T7-RXRα ΔAF1 and Flag-TGIF, as indicated. Complexes were collected on Flag agarose and analyzed for the presence of coprecipitating T7-tagged proteins. A portion of the lysates was analyzed by direct Western blotting (below). IP, immunoprecipitation; WB, Western blotting; +, present.
FIG. 6.
TGIF recruits CtBP to RXRα. (A) COS1 cells were transfected with expression plasmids encoding T7-tagged RXRα ΔAF1 and TGIF (amino acids 1 to 116) and Flag-tagged CtBP, as indicated. Proteins were precipitated on Flag agarose and analyzed for the presence of coprecipitating T7-RXRα ΔAF1. A portion of the lysates was analyzed by direct Western blotting (below). (B) HepG2 cells were transfected with a DR1-TATA-luc luciferase reporter, and expression vectors encoding RXRα and TGIF or a mutant form of TGIF (S28C), as indicated. 9C-RA was added for the indicated times prior to analysis. Luciferase activity is presented, in arbitrary units, as the mean ± standard deviation of the results from duplicate transfections. (C) COS1 cells were transfected with T7-RXRα ΔAF1 and a control vector or one encoding Flag-TGIF or the S28C mutant form of TGIF. Flag immunoprecipitates were analyzed for the presence of T7-RXRα ΔAF1, and expression of transfected proteins in the lysates is shown below. IP, immunoprecipitation; WB, Western blotting; +, present.
FIG. 7.
A Tgif null mutation in mice. (A) The targeting vector, Tgif locus, and targeted allele are shown, with restriction enzyme sites and predicted sizes of restriction fragments. Coding exons are shown in black, and noncoding exons are shown in gray. The arrows indicate the positions of PCR primers used for genotype analyses. The hatched gray bar indicates the position of the probe used for Southern blotting. (B) Genomic DNA from mice of the indicated genotypes was subjected to restriction enzyme digestion and Southern blotting. (C) RNA from mice of the indicated genotypes was subjected to Northern analysis with probes for Tgif, GFP, Tgif2, and Gapdh. (D) Heterozygous Tgif mutants were intercrossed, and the genotypes of the offspring at 21 days after birth were determined by PCR from genomic DNA. The positions of the PCR primers are indicated by arrows in panel A. The numbers of mice of each genotype and the percentage of the total are shown, together with the number of litters and average litter size.
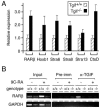
FIG. 8.
Analysis of endogenous gene expression in Tgif null cells. (A) RNA was isolated from wild-type or mutant testes and subjected to real time RT-PCR analysis. Expression of the indicated genes was analyzed in triplicate together with actin, and results are presented as expression relative to actin for the wild type and mutant, with expression of each gene arbitrarily set equal to 1 for the wild-type samples. (B) Wild-type or Tgif null mouse embryo fibroblasts were treated with (+) or without (−) RA for 16 h, as indicated. Chromatin was cross-linked with formaldehyde and subjected to immunoprecipitation with a TGIF-specific antiserum (α-TGIF) or the preimmune serum (Pre-imm). Precipitates were analyzed by PCR for the presence of a region of the RARβ promoter which spans the RARE or part of the GAPDH promoter. In addition, the input chromatin samples were analyzed with the same oligonucleotide pairs.
FIG. 9.
Retinoic acid teratogenesis in Tgif mutant embryos. Pregnant females, from heterozygous intercrosses, were treated with retinoic acid at gestational day 7.5. Embryos were examined for developmental defects at E10.5. (A) Embryos representative of each defect are shown. A normal wild type embryo and a homozygous mutant with exencephaly (severe open neural tube) are shown above. Below are three heterozygous mutants, one with a reduced fore and hind brain (left) and normal and arrested littermates (right). (B) The percentage of defective embryos (all defects) of each Tgif genotype is shown, together with the percentage with exencephaly. The total number of embryos of each genotype analyzed is shown below. (C) The numbers of embryos with each defect and the numbers of normal embryos are shown for each Tgif genotype. Defects: fb + hb, reduced fore and hind brain; arrest, severe developmental arrest or delay; exenc, exencephaly.
Similar articles
- Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice.
Shen J, Walsh CA. Shen J, et al. Mol Cell Biol. 2005 May;25(9):3639-47. doi: 10.1128/MCB.25.9.3639-3647.2005. Mol Cell Biol. 2005. PMID: 15831469 Free PMC article. - The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF.
Melhuish TA, Wotton D. Melhuish TA, et al. J Biol Chem. 2000 Dec 15;275(50):39762-6. doi: 10.1074/jbc.C000416200. J Biol Chem. 2000. PMID: 10995736 - Negative interplay of retinoic acid and TGF-β signaling mediated by TG-interacting factor to modulate mouse embryonic palate mesenchymal-cell proliferation.
Liu X, Zhang H, Gao L, Yin Y, Pan X, Li Z, Li N, Li H, Yu Z. Liu X, et al. Birth Defects Res B Dev Reprod Toxicol. 2014 Dec;101(6):403-9. doi: 10.1002/bdrb.21130. Epub 2014 Dec 4. Birth Defects Res B Dev Reprod Toxicol. 2014. PMID: 25477235 - TGIF2 interacts with histone deacetylase 1 and represses transcription.
Melhuish TA, Gallo CM, Wotton D. Melhuish TA, et al. J Biol Chem. 2001 Aug 24;276(34):32109-14. doi: 10.1074/jbc.M103377200. Epub 2001 Jun 26. J Biol Chem. 2001. PMID: 11427533 - Role of TG-interacting factor (Tgif) in lipid metabolism.
Pramfalk C, Eriksson M, Parini P. Pramfalk C, et al. Biochim Biophys Acta. 2015 Jan;1851(1):9-12. doi: 10.1016/j.bbalip.2014.07.019. Epub 2014 Aug 1. Biochim Biophys Acta. 2015. PMID: 25088698 Review.
Cited by
- Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation.
Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D. Powers SE, et al. Development. 2010 Jan;137(2):249-59. doi: 10.1242/dev.040782. Development. 2010. PMID: 20040491 Free PMC article. - Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development.
Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Hernandez RE, et al. Development. 2007 Jan;134(1):177-87. doi: 10.1242/dev.02706. Development. 2007. PMID: 17164423 Free PMC article. - Retinoic acid, GABA-ergic, and TGF-beta signaling systems are involved in human cleft palate fibroblast phenotype.
Baroni T, Bellucci C, Lilli C, Pezzetti F, Carinci F, Becchetti E, Carinci P, Stabellini G, Calvitti M, Lumare E, Bodo M. Baroni T, et al. Mol Med. 2006 Sep-Oct;12(9-10):237-45. doi: 10.2119/2006–00026.Baroni. Mol Med. 2006. PMID: 17225872 Free PMC article. - Cooperative transcriptional activation by Klf4, Meis2, and Pbx1.
Bjerke GA, Hyman-Walsh C, Wotton D. Bjerke GA, et al. Mol Cell Biol. 2011 Sep;31(18):3723-33. doi: 10.1128/MCB.01456-10. Epub 2011 Jul 11. Mol Cell Biol. 2011. PMID: 21746878 Free PMC article. - Retinoic acid drives intestine-specific adaptation of effector ILC2s originating from distant sites.
Shaikh N, Waterhölter A, Gnirck AC, Becker M, Adamiak V, Henneken L, Wunderlich M, Hartmann W, Linnemann L, Huber TB, Krebs CF, Panzer U, Locksley RM, Wilhelm C, Breloer M, Turner JE. Shaikh N, et al. J Exp Med. 2023 Dec 4;220(12):e20221015. doi: 10.1084/jem.20221015. Epub 2023 Sep 29. J Exp Med. 2023. PMID: 37773047 Free PMC article.
References
- Allenby, G., R. Janocha, S. Kazmer, J. Speck, J. F. Grippo, and A. A. Levin. 1994. Binding of 9-cis-retinoic acid and all-trans-retinoic acid to retinoic acid receptors alpha, beta, and gamma. Retinoic acid receptor gamma binds all-trans-retinoic acid preferentially over 9-cis-retinoic acid. J. Biol. Chem. 269:16689-16695. - PubMed
- Bastien, J., and C. Rochette-Egly. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1-16. - PubMed
- Bertolino, E., B. Reimund, D. Wildt-Perinic, and R. Clerc. 1995. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270:31178-31188. - PubMed
- Chambon, P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940-954. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous