Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA - PubMed (original) (raw)
Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA
Sujatha Kadaba et al. RNA. 2006 Mar.
Abstract
1-Methyladenosine modification at position 58 of tRNA is catalyzed by a two-subunit methyltransferase composed of Trm6p and Trm61p in Saccharomyces cerevisiae. Initiator tRNA (tRNAi(Met)) lacking m1A58 (hypomethylated) is rendered unstable through the cooperative function of the poly(A) polymerases, Trf4p/Trf5p, and the nuclear exosome. We provide evidence that a catalytically active Trf4p poly(A) polymerase is required for polyadenylation of hypomethylated tRNAi(Met) in vivo. DNA sequence analysis of tRNAi(Met) cDNAs and Northern hybridizations of poly(A)+ RNA provide evidence that nascent pre-tRNAi(Met) transcripts are targeted for polyadenylation and degradation. We determined that a mutant U6 snRNA and an aberrant form of 5S rRNA are stabilized in the absence of Trf4p, supporting that Trf4p facilitated RNA surveillance is a global process that stretches beyond hypomethylated tRNAi(Met). We conclude that an array of RNA polymerase III transcripts are targeted for Trf4p/ Trf5p-dependent polyadenylation and turnover to eliminate mutant and variant forms of normally stable RNAs.
Figures
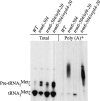
FIGURE 1.
Detection of polyadenylated tRNAiMet in trm6-504 and trm6-504 suppressor mutants. Total RNA and poly(A)+ RNA were isolated from wild-type (Y200), trm6-504(Y190), trm6-504 trf4-20 (sup1), and trm6-504 rrp44-20(sup2) strains grown in YPD at 30°C as detailed in Materials and Methods. Total RNA (5 μg) and poly(A)+ RNA (2–4 μg) were separated on a 6% polyacrylamide (19:1) 8 Murea gel. Membrane-bound tRNAiMet was detected using a radiolabeled oligonucleotide JA11 followed by autoradiography.
FIGURE 2.
The DXD motif of Trf4p is crucial for its poly(A) polymerase activity and the degradation of tRNAiMet. (A) Sctrf4-236 complements the cold sensitivity of trf4_Δ. A strain bearing chromosomal trf4_Δ was transformed with YCplac33, Sc_TRF4 (B254), or Sc_trf4-236 (B316), and the transformants were grown on SC-URA at 30°C, then replica-printed onto SC-URA plates and incubated at 16° and 26°C for 3 d and 1 d, respectively. (B) trf4-236 suppresses the Ts− phenotype of a trm6-504 mutant. A trm6-504 trf4_Δ strain (Y301) was transformed with YCpLac33, Sc_TRF4 (B254), and Sc_trf4-236_ (B316); wild type (Y200) and trm6-504 (Y190) were transformed just with YCpLac33. Individual transformants were grown to saturation in SC-URA and 10-fold serial dilutions were spotted on SC-URA plates and incubated at 26°, 36°, and 16°C for 2, 3, and 4 d, respectively. (C) Northern hybridization of total RNA (5 μg) from wild type (Y200), trm6-504(Y190), trm6-504 trf4_Δ (sup1) with YCpLac33, or Sc_trf4-236 strains. Total RNA (5 μg) was separated on a 6% polyacrylamide (19:1) 8 Murea gel and transferred to a membrane and probed with JA11 to detect tRNAiMet and JA 151 to detect tRNALeu. The tRNAs were visualized by autoradiography and the quantities were determined by phosphorimage analysis using a STORM 640 and ImageQuant software (GE Healthcare). The quantity of tRNAiMet in each sample is shown as a percentage of tRNAiMet in the wild-type strain after normalization with tRNACAALeu. This experiment has been repeated and the quantity of tRNAiMet was found to be similar each time. (D) The DXD motif is important for its poly(A) polymerase activity in vivo. Total RNA (5 μg) and poly(A)+ RNA (2–4 μg) isolated from the transformants described in B were separated, and a Northern hybridization was conducted to detect tRNAiMet using a radiolabeled oligonucleotide (JA11) and autoradiography.
FIGURE 3.
Trf4p and Trf5p have overlapping functions in the degradation of hypomethylated tRNAiMet. (A) Yeast strains wild type (Y200), trm6-504 (Y190), trm6-504 trf4-20 (sup1), and _trm6-504 trf5_Δ were grown to saturation in YPD and serial 10-fold dilutions were plated onto YPD plates and incubated at 26°C or 36°C for 3 d and 2 d, respectively. (B) The same strains from A transformed with empty vector (WT, trm6-504, and _trm6-504 trf5_Δ) or a high-copy number plasmid containing TRF5 (WT, trm6-504, and trm6-504 trf5_Δ + hc_TRF5) were streaked to synthetic complete media lacking uracil and incubated at 30°C and 36°C for 3 d. (C) Northern hybridization of total RNA (3 μg) and poly(A)+ RNA (4–5 μg) isolated from wild-type (Y200), _trm6-504 trf4_Δ and trm6-504 trf4_Δ + hc_TRF5. The blot was hybridized with a radiolabeled oligonucleotide (JA11) to detect tRNAiMet.
FIGURE 4.
Precursor form of hypomodified tRNAiMet is being polyadenylated. (A) An abbreviated and stylized representation of the tRNAiMet coding sequence with the predicted 3′ ends of each pre-tRNAiMet synthesized from IMT1–4 shown. The predicted transcription termination sites of pre-tRNAiMet are shown as 5–7 tandem Ts (Us in the transcript) are shown in bold. (B) The abbreviated coding sequence is followed by the exact sequence found at the 3′ end of each cDNA, and the poly(A) tail lengths are shown as a range (16–30), but the majority of the poly(A) tails were 16–19 As in length. The number of clones with the identical 3′ end sequence are shown in parentheses. (C) The abbreviated coding sequence for the truncated cDNAs found end with the last nucleotide of the tRNA (C65 or C60) followed by the poly(A) tail length for each.
FIGURE 5.
Polyadenylated pre-tRNAiMet contains the 5′ leader sequence. (A) Northern hybridization analysis of total RNA (5 μg) and poly(A)+ RNA (2–4 μg) from trm6-504 (Y190) and wild type (Y200). (B) Northern hybridization of total RNA (5 μg) and poly(A)+ RNA (2–4 μg) from _trm6-504 rrp6_Δ (Y298) with (+) or without (−) pretreatment with oligo d(T) and RNaseH. The blots were hybridized with radiolabeled probes (JA382, JA72, JA66, and JA48) that recognize only pre-tRNAiMet containing the 5′ leader (A,B, lanes 1–3) or a radiolabeled probe (JA11) that is complementary to an internal portion of tRNAiMet (B, lanes 4–6). The point of migration of pre-tRNAiMet and mature tRNAiMet are shown for clarity.
FIGURE 6.
Internally truncated U6 snRNA is a substrate for Trf4p polyadenylation and degradation by the nuclear exosome. Wild type (F39), _rrp6_Δ (F23), _trf4_Δ (F22), and trf4_Δ + Sc_trf4-236 were transformed with a single-copy number plasmid containing a mutant SNR6 gene (snr6_Δ_59–72). Total RNA (5 μg) from single transformants and a wildtype strain bearing an empty plasmid were analyzed by Northern hybridization using a radiolabeled probe (JA242) that recognizes the endogenous and plasmid-borne U6 snRNAs. The migration positions of the endogenous and snr6Δ59–72 mutant U6 snRNAs are shown for clarity.
FIGURE 7.
Trf4p dependent polyadenylation of 3′ end truncated 5S rRNA. (A) A schematic diagram of 5S rDNA showing the coding region (filled box) and the 5′ and 3′ intergenic regions (lines). The approximate positions (not to scale) complementary to the oligonucleotide probes used in this study are shown as lines beneath the coding region. (B) Total RNA (5 μg) isolated from wild type (F39), _rrp6_Δ (F23), rrp44-20 (Y303), _trf4_Δ (F22), and _trm6-504/trf4_Δ (Y301) strains grown at 30°C was used for Northern hybridization analysis. The membrane was probed with JA99 to detect mature 5S rRNA, JA133 to detect 5S rRNA containing the first 20 nt of 5S rRNA and JA134 to detect 5S rRNA containing the last 20 nt. The 5S rRNA was visualized by autoradiography. (C) Total RNA (5 μg, lanes 1–5) and poly(A)+ RNA (2–4 μg, lanes 6–10) isolated from the same strains described in B was used for Northern hybridization analysis using probes JA133 (top) and JA134 (bottom). Indicated are the migration positions of the mature 5S rRNA, 3′-end truncated 5S rRNA, and poly(A)+ 5S rRNA, as well as the probe used for each hybridization.
Similar articles
- Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome.
Egecioglu DE, Henras AK, Chanfreau GF. Egecioglu DE, et al. RNA. 2006 Jan;12(1):26-32. doi: 10.1261/rna.2207206. RNA. 2006. PMID: 16373491 Free PMC article. - Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae.
Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Kadaba S, et al. Genes Dev. 2004 Jun 1;18(11):1227-40. doi: 10.1101/gad.1183804. Epub 2004 May 14. Genes Dev. 2004. PMID: 15145828 Free PMC article. - Distinct roles of non-canonical poly(A) polymerases in RNA metabolism.
San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP. San Paolo S, et al. PLoS Genet. 2009 Jul;5(7):e1000555. doi: 10.1371/journal.pgen.1000555. Epub 2009 Jul 10. PLoS Genet. 2009. PMID: 19593367 Free PMC article. - The impact of transcription on posttranscriptional processes in yeast.
Turowski TW. Turowski TW. Gene. 2013 Aug 15;526(1):23-9. doi: 10.1016/j.gene.2013.04.021. Epub 2013 Apr 30. Gene. 2013. PMID: 23639960 Review. - Use of mutant RNAs in studies on yeast 5S rRNA structure and function.
Nazar RN, Van Ryk DI, Lee Y, Guyer CD. Nazar RN, et al. Biochem Cell Biol. 1991 Apr;69(4):217-22. doi: 10.1139/o91-033. Biochem Cell Biol. 1991. PMID: 2054154 Review.
Cited by
- Advances in the Structural and Functional Understanding of m1A RNA Modification.
Smoczynski J, Yared MJ, Meynier V, Barraud P, Tisné C. Smoczynski J, et al. Acc Chem Res. 2024 Feb 8;57(4):429-38. doi: 10.1021/acs.accounts.3c00568. Online ahead of print. Acc Chem Res. 2024. PMID: 38331425 Free PMC article. - Transcriptome-wide analysis of exosome targets.
Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Schneider C, et al. Mol Cell. 2012 Nov 9;48(3):422-33. doi: 10.1016/j.molcel.2012.08.013. Epub 2012 Sep 20. Mol Cell. 2012. PMID: 23000172 Free PMC article. - The exosome and RNA quality control in the nucleus.
Vanacova S, Stefl R. Vanacova S, et al. EMBO Rep. 2007 Jul;8(7):651-7. doi: 10.1038/sj.embor.7401005. EMBO Rep. 2007. PMID: 17603538 Free PMC article. Review. - Mutations in the anticodon stem of tRNA cause accumulation and Met22-dependent decay of pre-tRNA in yeast.
Payea MJ, Hauke AC, De Zoysa T, Phizicky EM. Payea MJ, et al. RNA. 2020 Jan;26(1):29-43. doi: 10.1261/rna.073155.119. Epub 2019 Oct 16. RNA. 2020. PMID: 31619505 Free PMC article. - A comprehensive analysis of the La-motif protein superfamily.
Bousquet-Antonelli C, Deragon JM. Bousquet-Antonelli C, et al. RNA. 2009 May;15(5):750-64. doi: 10.1261/rna.1478709. Epub 2009 Mar 19. RNA. 2009. PMID: 19299548 Free PMC article.
References
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., and Hinnebusch, A.G. 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 12: 3650–3662. - PMC - PubMed
- Arnez, J.G. and Moras, D. 1999. Transfer RNA. In Oxford handbook of nucleic acid structure (ed. S. Neidle), pp. 603–651. Oxford University Press, Oxford, UK.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous